

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093a.1
(268 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
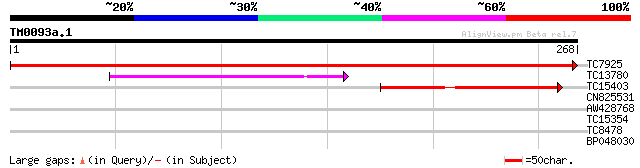
TC7925 homologue to UP|RPE_SPIOL (Q43157) Ribulose-phosphate 3-e... 526 e-150
TC13780 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-e... 91 2e-19
TC15403 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-e... 69 6e-13
CN825531 33 0.066
AW428768 30 0.33
TC15354 similar to PIR|B86314|B86314 F2H15.12 protein - Arabidop... 29 0.73
TC8478 similar to PIR|B86367|B86367 protein F26F24.16 [imported]... 28 1.2
BP048030 28 2.1
>TC7925 homologue to UP|RPE_SPIOL (Q43157) Ribulose-phosphate 3-epimerase,
chloroplast precursor (Pentose-5-phosphate 3-epimerase)
(PPE) (RPE) (R5P3E) , partial (94%)
Length = 1387
Score = 526 bits (1354), Expect = e-150
Identities = 268/268 (100%), Positives = 268/268 (100%)
Frame = +3
Query: 1 INGPSSLHSKTAIFNNHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKL 60
INGPSSLHSKTAIFNNHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKL
Sbjct: 189 INGPSSLHSKTAIFNNHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKL 368
Query: 61 GEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVP 120
GEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVP
Sbjct: 369 GEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVP 548
Query: 121 DFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL 180
DFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL
Sbjct: 549 DFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL 728
Query: 181 IMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALV 240
IMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALV
Sbjct: 729 IMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALV 908
Query: 241 AGSAVFGAKDYAEAIKGIKTSKRPEPVA 268
AGSAVFGAKDYAEAIKGIKTSKRPEPVA
Sbjct: 909 AGSAVFGAKDYAEAIKGIKTSKRPEPVA 992
>TC13780 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-epimerase ,
partial (50%)
Length = 445
Score = 91.3 bits (225), Expect = 2e-19
Identities = 46/113 (40%), Positives = 68/113 (59%)
Frame = +1
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PS+LS++FA L + + + G DW+H+D+MDG FVPN+TIG ++++LR T LD
Sbjct: 106 IAPSMLSSDFANLASEAQRMLDYGADWLHMDIMDGHFVPNLTIGTPVIESLRKHTKAYLD 285
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVL 160
HLM+ P V KAGA + H E S + + ++KS G K GV L
Sbjct: 286 CHLMVTNPLDYVEPLGKAGASGFTFHVEASKD-NWKEVIQRIKSQGMKPGVAL 441
>TC15403 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-epimerase ,
partial (40%)
Length = 562
Score = 69.3 bits (168), Expect = 6e-13
Identities = 38/86 (44%), Positives = 55/86 (63%)
Frame = +2
Query: 176 VDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAG 235
V++VL+M+V PGFGGQ F+ + K+ LR+ K + IEVDGG+ P+ AG
Sbjct: 23 VEMVLVMTVAPGFGGQKFMPEMMDKVRILRK----KYPSLDIEVDGGLGPSTIDMAASAG 190
Query: 236 ANALVAGSAVFGAKDYAEAIKGIKTS 261
AN +VAGS+VFGA + A+ I ++ S
Sbjct: 191 ANCIVAGSSVFGAPEPAQVISLLRNS 268
>CN825531
Length = 627
Score = 32.7 bits (73), Expect = 0.066
Identities = 24/82 (29%), Positives = 37/82 (44%)
Frame = -2
Query: 136 QSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIE 195
++ST L TV+Q+ A + +PG PL I + + D I ++ GFG
Sbjct: 200 KASTKSLQETVSQLG*ALCAASSLYDPGIPL*DIRTLAPLSDAATISDMDSGFG------ 39
Query: 196 SQVKKISDLRRLCAEKGVNPWI 217
S+L+ LC NPW+
Sbjct: 38 ------SELKSLC-----NPWV 6
>AW428768
Length = 286
Score = 30.4 bits (67), Expect = 0.33
Identities = 14/23 (60%), Positives = 17/23 (73%)
Frame = +3
Query: 193 FIESQVKKISDLRRLCAEKGVNP 215
+IESQ+ KI LR+LCAEK P
Sbjct: 123 YIESQLNKIYLLRKLCAEKATVP 191
>TC15354 similar to PIR|B86314|B86314 F2H15.12 protein - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (43%)
Length = 668
Score = 29.3 bits (64), Expect = 0.73
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Frame = +3
Query: 131 SIHCEQSSTIH-LHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIM 182
+ H E S + L R ++ K GAK VV G+PL +V D+ D V+ M
Sbjct: 18 NFHPENSHVLPALMRRFHEAKVKGAKEVVVWGTGSPLREFLHVDDLADAVVFM 176
>TC8478 similar to PIR|B86367|B86367 protein F26F24.16 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(67%)
Length = 1124
Score = 28.5 bits (62), Expect = 1.2
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 7/47 (14%)
Frame = +3
Query: 126 GADIVSIHCEQSS-----TIHLHRTVNQV--KSLGAKAGVVLNPGTP 165
G +V + E+++ T L R+V Q K + KA V+LNPG P
Sbjct: 672 GGTLVGYYLEETANWGLDTKELRRSVEQARYKGINVKAMVILNPGNP 812
>BP048030
Length = 323
Score = 27.7 bits (60), Expect = 2.1
Identities = 15/39 (38%), Positives = 21/39 (53%)
Frame = -3
Query: 144 RTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIM 182
R ++ K GAK VV G+PL +V D+ D V+ M
Sbjct: 318 RRFHEAKVKGAKEVVVWGTGSPLREFLHVDDLADAVVFM 202
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,822,251
Number of Sequences: 28460
Number of extensions: 42632
Number of successful extensions: 217
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 216
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 216
length of query: 268
length of database: 4,897,600
effective HSP length: 89
effective length of query: 179
effective length of database: 2,364,660
effective search space: 423274140
effective search space used: 423274140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0093a.1


