

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.2
(153 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
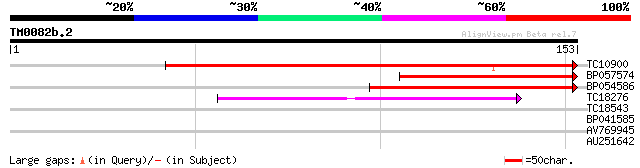
Score E
Sequences producing significant alignments: (bits) Value
TC10900 weakly similar to UP|Q9CB03 (Q9CB03) F17L21.25 (AT1G2746... 172 2e-44
BP057574 62 3e-11
BP054586 59 3e-10
TC18276 similar to UP|O82039 (O82039) SPINDLY protein, partial (... 38 9e-04
TC18543 similar to UP|Q94EJ1 (Q94EJ1) At1g22700/T22J18_13, parti... 28 0.54
BP041585 27 1.2
AV769945 27 1.6
AU251642 26 3.5
>TC10900 weakly similar to UP|Q9CB03 (Q9CB03) F17L21.25
(AT1G27460/F17L21_26), partial (5%)
Length = 609
Score = 172 bits (437), Expect = 2e-44
Identities = 85/114 (74%), Positives = 99/114 (86%), Gaps = 3/114 (2%)
Frame = +2
Query: 43 YTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRS 102
Y+ASRCH G M+EAKGLYKEA+ AFR+AL+IDP HVPSLISTA+ L+R +N S+P VRS
Sbjct: 5 YSASRCHASGMMYEAKGLYKEALTAFREALDIDPDHVPSLISTALVLKRCTNPSNPAVRS 184
Query: 103 FLMDALRYDRLNASAWYNLGILHKAEG---RVLEAVECFQAANSLEETAPIEPF 153
FLMDALR+DRLNASAWYNLG+LHKAEG V+EA ECFQAA SLEE+AP+EPF
Sbjct: 185 FLMDALRHDRLNASAWYNLGLLHKAEGTASSVVEAAECFQAAYSLEESAPVEPF 346
>BP057574
Length = 507
Score = 62.4 bits (150), Expect = 3e-11
Identities = 29/48 (60%), Positives = 35/48 (72%)
Frame = -2
Query: 106 DALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETAPIEPF 153
DALR + N AWY LG+ HKA+GRV +A +CFQAA LEE+ PIE F
Sbjct: 503 DALRIEPTNRMAWYYLGLTHKADGRVGDAADCFQAAYMLEESEPIEDF 360
>BP054586
Length = 340
Score = 59.3 bits (142), Expect = 3e-10
Identities = 26/56 (46%), Positives = 40/56 (71%)
Frame = -1
Query: 98 PVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETAPIEPF 153
P+ RSFLM+ALR + N W+NLG++ K+EG + +A + FQAA L+ +AP++ F
Sbjct: 310 PIARSFLMNALRIEPENHDVWFNLGLVSKSEGSLQQAADYFQAAYELKLSAPVQKF 143
>TC18276 similar to UP|O82039 (O82039) SPINDLY protein, partial (10%)
Length = 660
Score = 37.7 bits (86), Expect = 9e-04
Identities = 22/82 (26%), Positives = 42/82 (50%)
Frame = +1
Query: 57 AKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNAS 116
++ + +++ + L D +V +LI + L+ + L +A++ D NA
Sbjct: 421 SRNKFVDSLAMYERVLETDNGNVEALIGKGICLQM--QNMGRLAFDSLSEAIKLDPQNAC 594
Query: 117 AWYNLGILHKAEGRVLEAVECF 138
A + GIL+K EGR++EA E +
Sbjct: 595 ALTHCGILYKEEGRLMEAAESY 660
>TC18543 similar to UP|Q94EJ1 (Q94EJ1) At1g22700/T22J18_13, partial (41%)
Length = 518
Score = 28.5 bits (62), Expect = 0.54
Identities = 12/28 (42%), Positives = 18/28 (63%)
Frame = +1
Query: 51 IGTMHEAKGLYKEAVKAFRDALNIDPRH 78
+G +E+K YK A+KAF + L DP +
Sbjct: 220 LGDAYESKKDYKSALKAFEEVLLFDPHN 303
>BP041585
Length = 503
Score = 27.3 bits (59), Expect = 1.2
Identities = 14/43 (32%), Positives = 21/43 (48%)
Frame = +1
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVE 136
N++ P SF L NA+ WYN+G + E R + V+
Sbjct: 67 NRTVPGATSFPAVILSAASFNATLWYNMGQVVSTEARAMYNVD 195
>AV769945
Length = 284
Score = 26.9 bits (58), Expect = 1.6
Identities = 13/27 (48%), Positives = 18/27 (66%)
Frame = -1
Query: 115 ASAWYNLGILHKAEGRVLEAVECFQAA 141
A A+ NLG++ AEG + EA CF+ A
Sbjct: 239 APAFNNLGLVFVAEGLLEEAKYCFEKA 159
>AU251642
Length = 427
Score = 25.8 bits (55), Expect = 3.5
Identities = 9/13 (69%), Positives = 12/13 (92%)
Frame = -2
Query: 16 DLAYAYISLSQWH 28
+L+YAYI+L QWH
Sbjct: 342 NLSYAYIALLQWH 304
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,584,102
Number of Sequences: 28460
Number of extensions: 29261
Number of successful extensions: 129
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 128
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 128
length of query: 153
length of database: 4,897,600
effective HSP length: 82
effective length of query: 71
effective length of database: 2,563,880
effective search space: 182035480
effective search space used: 182035480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0082b.2


