

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.15
(1211 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
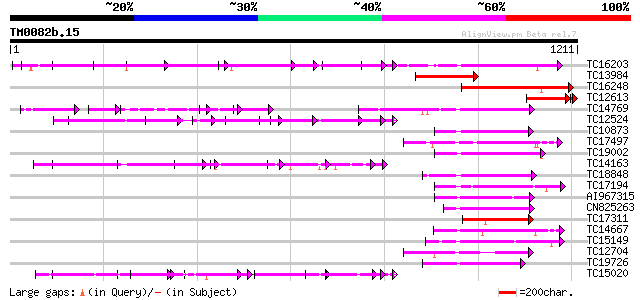
Score E
Sequences producing significant alignments: (bits) Value
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 410 e-115
TC13984 weakly similar to UP|Q9LVP0 (Q9LVP0) Receptor-like prote... 275 2e-74
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 233 1e-61
TC12613 180 5e-49
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 165 5e-41
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 164 6e-41
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 156 2e-38
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 154 1e-37
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 149 2e-36
TC14163 149 3e-36
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 147 1e-35
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 142 2e-34
AI967315 136 2e-32
CN825263 135 5e-32
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 134 9e-32
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 132 3e-31
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 132 3e-31
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 130 9e-31
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 130 9e-31
TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-... 130 1e-30
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 410 bits (1055), Expect = e-115
Identities = 293/953 (30%), Positives = 464/953 (47%), Gaps = 21/953 (2%)
Frame = +2
Query: 249 GPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDFQGPIPSSIGNLTM 308
G +P EI L L+ L + +NNL +P ++ L+ L+VL + N F G P GN+T+
Sbjct: 371 GHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFP---GNITV 541
Query: 309 LQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLSMASLTRMRELGLSSNQLS 368
G+ T L +D NS +G LP + L +++ L L+ N S
Sbjct: 542 ---------GM-----------TELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFS 661
Query: 369 GELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHLYL-YENQFSGPIPKEIGNL 427
G + P S + L L L N +TG++P + L L+L Y N + G IP G++
Sbjct: 662 GTI-PESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSM 838
Query: 428 SSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLSGALPPEIGDLENLEELKLSENH 487
++ L+++N + G IP ++G L KL +L + N L+G +PPE+ + +L L LS N
Sbjct: 839 ENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSIND 1018
Query: 488 LSGPLPSSITHLENIKILHLHWNNFSGSIPEDFG--PNFLTNVSFANNSFSGNLPSGICR 545
L+G +P S + L+N+ +++ N F GS+P G PN T + NN FS LP +
Sbjct: 1019 LTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENN-FSFVLPHNLGG 1195
Query: 546 GGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNLLSGDITNAFGTYPDLNFIDLGHN 605
G +Y N+ G IP L L ++ +N G I G
Sbjct: 1196 NGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIG------------- 1336
Query: 606 QLSGSLSSNWGECKFLSSFSISSNKVHGNIPPELGKLRLQNL-DLSENNLTGNIPVELFN 664
EC+ L+ +++N + G +PP + +L + +LS N L G +P +
Sbjct: 1337 -----------ECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVI-- 1477
Query: 665 SSSKMLKLNLSNNYLSGHMPTRIGELSELQYLDFSANNLSGPIPNALGNCGNLIFLKLSM 724
S + L LSNN +G +P + L LQ L AN G IP + L + +S
Sbjct: 1478 SGESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISG 1657
Query: 725 NNLEGPMPHELGNLVNLQPLLDLSHNSLSGAIIPQLEKLTSLEVLNLSHNQLSGGIPSDL 784
NNL GP+P + + +L + DLS N+L+G + ++ L L +LNLS N++SG +P ++
Sbjct: 1658 NNLTGPIPTTITHRASLTAV-DLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEI 1834
Query: 785 NGLISLQSIDISYNKLEGPLPSLEAFHNAS-EEALVGNSGLCSGPDNGNANLSPCGGEKS 843
+ SL ++D+S N G +P+ F + ++ GN LC P + +
Sbjct: 1835 RFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCF-PHRASCPSVLYDSLRK 2011
Query: 844 NKDNNHKLIIAIVIPVA-ALIILLVSLGLFFIFRRYRKAGKAKKDKGSNRKNSFFIWNHR 902
+ + + AIVI +A A +LLV++ + + +R +A K R
Sbjct: 2012 TRAKTAR-VRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAFQRLE-------- 2164
Query: 903 NRIEFEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRL-HQNEEMEDFPGYQA 961
I+ ED+ E E+ IG GG G VY+ +P G A+KRL Q D+
Sbjct: 2165 --IKAEDV---VECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDY----- 2314
Query: 962 KNFTSEIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQKEQEAKILTWDI 1021
F +EI L IRHRN++++ G+ + +YE++ GSL L + L W++
Sbjct: 2315 -GFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH-LRWEM 2488
Query: 1022 RLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLL---KAGAN 1078
R + A L Y+HHDC+P I+HRD+ NN+LLD+++E ++DFG A+ L A +
Sbjct: 2489 RYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQS 2668
Query: 1079 WTTPVGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYPHEVL---------LCLE 1129
++ GSYGY+APE A T+KV EK DVYSFGVV L++++G+ P +
Sbjct: 2669 MSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKT 2848
Query: 1130 SRELDQHFIDFLDKRPTPP--EGPAIQLLVMVATLILKCVAKDPLSRPTMRQV 1180
EL Q L P G + ++ + + + CV + +RPTMR+V
Sbjct: 2849 MSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREV 3007
Score = 263 bits (673), Expect = 9e-71
Identities = 157/507 (30%), Positives = 253/507 (48%), Gaps = 27/507 (5%)
Frame = +2
Query: 180 PDQFKGMKSMTELNLSYNSLTD-VPPFVSKCPKLVSLDLSLNTITGKIPIHLLTNLKNLT 238
P + ++ + L +S N+LTD +P ++ L L++S N +G+ P ++ + L
Sbjct: 380 PPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELE 559
Query: 239 ILDLTENRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDFQGP 298
LD +N F GP+PEEI L LK L L N +GTIP+ LE L L+ N G
Sbjct: 560 ALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGR 739
Query: 299 IPSSIGNLTMLQRLHLRLS-GLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLSMASLTRM 357
+P S+ L L+ LHL S IP G NL ++MA +LTG +P S+ +LT++
Sbjct: 740 VPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKL 919
Query: 358 RELGLSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHLYLYENQFS 417
L + N L+G + P LSS L+SL L +ND+TG++P NLT + ++N+F
Sbjct: 920 HSLFVQMNNLTGTIPPE-LSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFR 1096
Query: 418 GPIPKEIGNLSSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLSGALPPE------ 471
G +P IG+L ++ LQ+ N+F+ +P +G + + + N L+G +PP+
Sbjct: 1097GSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGR 1276
Query: 472 ------------------IGDLENLEELKLSENHLSGPLPSSITHLENIKILHLHWNNFS 513
IG+ +L +++++ N L GP+P + L ++ I L N +
Sbjct: 1277LKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLN 1456
Query: 514 GSIPEDFGPNFLTNVSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGL 573
G +P L ++ +NN F+G +P+ + L L+ + N F G IP + L
Sbjct: 1457GELPSVISGESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPML 1636
Query: 574 IRVLLGNNLLSGDITNAFGTYPDLNFIDLGHNQLSGSLSSNWGECKFLSSFSISSNKVHG 633
+V + N L+G I L +DL N L+G + LS ++S N++ G
Sbjct: 1637TKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISG 1816
Query: 634 NIPPELGKL-RLQNLDLSENNLTGNIP 659
+P E+ + L LDLS NN TG +P
Sbjct: 1817PVPDEIRFMTSLTTLDLSSNNFTGTVP 1897
Score = 242 bits (618), Expect = 2e-64
Identities = 170/613 (27%), Positives = 283/613 (45%), Gaps = 8/613 (1%)
Frame = +2
Query: 7 IAIFSLVLGCMLVAISAQKEAESLITWMNSLNSPLPS-----SWKLAGNNTSPCKWTSIS 61
+ F+L+ V S+ + ++L+ S+ WK + + ++ C ++ ++
Sbjct: 137 VLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSGVT 316
Query: 62 CDKAGTVVEIKLPNAGLDGTLNRFDFSAFPNLSNFNVSMNNLVGEIPSGIGNATKLKTLD 121
CD+ VV + + L G L + L N +SMNNL ++PS + + T LK L+
Sbjct: 317 CDQNLRVVALNVTLVPLFGHLPP-EIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLN 493
Query: 122 LGSNNLTNPIPPQIG-NLLELQVLIFSNNSLLKQIPSQLSNLQNLWLLDLGANYLENPDP 180
+ N + P I + EL+ L +NS +P ++ L+ L L L NY P
Sbjct: 494 ISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIP 673
Query: 181 DQFKGMKSMTELNLSYNSLTDVPPFVSKCPKLVSLDLSLNTITGKIPIHLLTNLKNLTIL 240
+ + +S+ L L+ NSL TG++P L LK L L
Sbjct: 674 ESYSEFQSLEFLGLNANSL-----------------------TGRVP-ESLAKLKTLKEL 781
Query: 241 DL-TENRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDFQGPI 299
L N +EG IP ++ NL+ L++ NL G IP +G+L+ L L + N+ G I
Sbjct: 782 HLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTI 961
Query: 300 PSSIGNLTMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLSMASLTRMRE 359
P + ++ L L L ++ L IP NL ++ N GSLP + L +
Sbjct: 962 PPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLET 1141
Query: 360 LGLSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHLYLYENQFSGP 419
L + N S L P L + + N +TG +PP + L + +N F GP
Sbjct: 1142LQVWENNFSFVL-PHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGP 1318
Query: 420 IPKEIGNLSSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLSGALPPEIGDLENLE 479
IPK IG S+ ++++NN +G +P + QL + L +N+L+G LP I E+L
Sbjct: 1319IPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISG-ESLG 1495
Query: 480 ELKLSENHLSGPLPSSITHLENIKILHLHWNNFSGSIPED-FGPNFLTNVSFANNSFSGN 538
L LS N +G +P+++ +L ++ L L N F G IP F LT V+ + N+ +G
Sbjct: 1496TLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGP 1675
Query: 539 LPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNLLSGDITNAFGTYPDLN 598
+P+ I +L + + NN G +P+ ++N L + L N +SG + + L
Sbjct: 1676IPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLT 1855
Query: 599 FIDLGHNQLSGSL 611
+DL N +G++
Sbjct: 1856TLDLSSNNFTGTV 1894
Score = 180 bits (457), Expect = 1e-45
Identities = 133/455 (29%), Positives = 211/455 (46%), Gaps = 11/455 (2%)
Frame = +2
Query: 26 EAESLITWMNSLNSPLPSS---------WKLAGNNTSPCKWTSISCDKAGTVVEIKLPNA 76
E E+L + NS + PLP LAGN S S S + ++ + L
Sbjct: 551 ELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYS--EFQSLEFLGLNAN 724
Query: 77 GLDGTLNRFDFSAFPNLSNFNVSMNNLV-GEIPSGIGNATKLKTLDLGSNNLTNPIPPQI 135
L G + + L ++ +N G IP G+ L+ L++ + NLT IPP +
Sbjct: 725 SLTGRVPE-SLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSL 901
Query: 136 GNLLELQVLIFSNNSLLKQIPSQLSNLQNLWLLDLGANYLENPDPDQFKGMKSMTELNLS 195
GNL +L L N+L IP +LS++ +L LDL N L P+ F +K++T +N
Sbjct: 902 GNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFF 1081
Query: 196 YNSLT-DVPPFVSKCPKLVSLDLSLNTITGKIPIHLLTNLKNLTILDLTENRFEGPIPEE 254
N +P F+ P L +L + N + +P H L D+T+N G IP +
Sbjct: 1082QNKFRGSLPSFIGDLPNLETLQVWENNFSFVLP-HNLGGNGRFLYFDVTKNHLTGLIPPD 1258
Query: 255 IKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDFQGPIPSSIGNLTMLQRLHL 314
+ LK + N G IP IG L + + N GP+P + L + L
Sbjct: 1259LCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITEL 1438
Query: 315 RLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLSMASLTRMRELGLSSNQLSGELYPS 374
+ LN +P+ I +L + ++ N TG +P +M +L ++ L L +N+ GE+ P
Sbjct: 1439SNNRLNGELPSVIS-GESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEI-PG 1612
Query: 375 LLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHLYLYENQFSGPIPKEIGNLSSINDLQ 434
+ P L + + N++TG +P I +LT + L N +G +PK + NL ++ L
Sbjct: 1613GVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILN 1792
Query: 435 LSNNHFNGSIPSTIGQLKKLITLALDSNQLSGALP 469
LS N +G +P I + L TL L SN +G +P
Sbjct: 1793LSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVP 1897
Score = 179 bits (453), Expect = 3e-45
Identities = 125/363 (34%), Positives = 185/363 (50%), Gaps = 4/363 (1%)
Frame = +2
Query: 447 TIGQLKKLITLALDSNQLSGALPPEIGDLENLEELKLSENHLSGPLPSSITHLENIKILH 506
T Q +++ L + L G LPPEIG LE LE L +S N+L+ LPS + L ++K+L+
Sbjct: 314 TCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLN 493
Query: 507 LHWNNFSGSIPEDF--GPNFLTNVSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIP 564
+ N FSG P + G L + +NSFSG LP I + L YL N F G IP
Sbjct: 494 ISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIP 673
Query: 565 ESLRNCTGLIRVLLGNNLLSGDITNAFGTYPDLNFIDLGH-NQLSGSLSSNWGECKFLSS 623
ES L + L N L+G + + L + LG+ N G + +G + L
Sbjct: 674 ESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRL 853
Query: 624 FSISSNKVHGNIPPELGKL-RLQNLDLSENNLTGNIPVELFNSSSKMLKLNLSNNYLSGH 682
+++ + G IPP LG L +L +L + NNLTG IP EL +S ++ L+LS N L+G
Sbjct: 854 LEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPEL-SSMMSLMSLDLSINDLTGE 1030
Query: 683 MPTRIGELSELQYLDFSANNLSGPIPNALGNCGNLIFLKLSMNNLEGPMPHELGNLVNLQ 742
+P +L L ++F N G +P+ +G+ NL L++ NN +PH LG
Sbjct: 1031IPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFL 1210
Query: 743 PLLDLSHNSLSGAIIPQLEKLTSLEVLNLSHNQLSGGIPSDLNGLISLQSIDISYNKLEG 802
D++ N L+G I P L K L+ ++ N G IP + SL I ++ N L+G
Sbjct: 1211-YFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDG 1387
Query: 803 PLP 805
P+P
Sbjct: 1388PVP 1396
Score = 112 bits (281), Expect = 3e-25
Identities = 82/275 (29%), Positives = 118/275 (42%), Gaps = 24/275 (8%)
Frame = +2
Query: 91 PNLSNFNVSMNNLVGEIPSGIGNATKLKTLDLGSNNLTNPIPPQIGNLLELQVLIFSNNS 150
PNL V NN +P +G + D+ N+LT IPP + L+ I ++N
Sbjct: 1127 PNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNF 1306
Query: 151 LLKQIPSQLSNLQNLWLLDLGANYLENPDPDQFKGMKSMTELNLSYNSLTDVPPFVSKCP 210
IP + ++L + + N+L+ P P + S+T LS N L P V
Sbjct: 1307 FRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGE 1486
Query: 211 KLVSLDLSLNTITGKIPIHLLTNLKNLTILDLTENRF----------------------- 247
L +L LS N TGKIP + NL+ L L L N F
Sbjct: 1487 SLGTLTLSNNLFTGKIPA-AMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNN 1663
Query: 248 -EGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDFQGPIPSSIGNL 306
GPIP I + ++L + L NNL G +P + +L L +L L +N+ GP+P I +
Sbjct: 1664 LTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFM 1843
Query: 307 TMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGN 341
T L L L + ++P G F Y AGN
Sbjct: 1844 TSLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGN 1948
Score = 76.3 bits (186), Expect = 3e-14
Identities = 50/160 (31%), Positives = 80/160 (49%)
Frame = +2
Query: 668 KMLKLNLSNNYLSGHMPTRIGELSELQYLDFSANNLSGPIPNALGNCGNLIFLKLSMNNL 727
+++ LN++ L GH+P IG L +L+ L S NNL+ +P+ L + +L L +S N
Sbjct: 332 RVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLF 511
Query: 728 EGPMPHELGNLVNLQPLLDLSHNSLSGAIIPQLEKLTSLEVLNLSHNQLSGGIPSDLNGL 787
G P + + LD NS SG + ++ KL L+ L+L+ N SG IP +
Sbjct: 512 SGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEF 691
Query: 788 ISLQSIDISYNKLEGPLPSLEAFHNASEEALVGNSGLCSG 827
SL+ + ++ N L G +P A +E +G S G
Sbjct: 692 QSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEG 811
Score = 44.7 bits (104), Expect = 9e-05
Identities = 28/77 (36%), Positives = 38/77 (48%)
Frame = +2
Query: 752 LSGAIIPQLEKLTSLEVLNLSHNQLSGGIPSDLNGLISLQSIDISYNKLEGPLPSLEAFH 811
L G + P++ L LE L +S N L+ +PSDL L SL+ ++IS+N G P
Sbjct: 365 LFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVG 544
Query: 812 NASEEALVGNSGLCSGP 828
EAL SGP
Sbjct: 545 MTELEALDAYDNSFSGP 595
>TC13984 weakly similar to UP|Q9LVP0 (Q9LVP0) Receptor-like protein kinase,
partial (4%)
Length = 404
Score = 275 bits (704), Expect = 2e-74
Identities = 133/134 (99%), Positives = 133/134 (99%)
Frame = +1
Query: 867 VSLGLFFIFRRYRKAGKAKKDKGSNRKNSFFIWNHRNRIEFEDICTATENFSEKYSIGTG 926
VSLGLFFIFRRYRKAGKAKKDKGSNRKNSFFIWNHRNRIEFEDICTATENFSEKYSIGTG
Sbjct: 1 VSLGLFFIFRRYRKAGKAKKDKGSNRKNSFFIWNHRNRIEFEDICTATENFSEKYSIGTG 180
Query: 927 GQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFS 986
GQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFS
Sbjct: 181 GQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFS 360
Query: 987 YLNGSMFFIYEHVE 1000
YLNGSM FIYEHVE
Sbjct: 361 YLNGSMCFIYEHVE 402
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 233 bits (594), Expect = 1e-61
Identities = 122/246 (49%), Positives = 167/246 (67%), Gaps = 7/246 (2%)
Frame = +2
Query: 966 SEIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQKEQEAKILTWDIRLNM 1025
SEI LT IRHRNVIK++GF + F +YE +E GSL++VL + +A W+ R+N+
Sbjct: 2 SEIQTLTEIRHRNVIKLHGFCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKRVNV 181
Query: 1026 IKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLLKAGAN-WTTPVG 1084
+KG+ANALSY+HHDC+P I+HRDIS NVLLD +YE +SDFGTA+ LK G++ WT G
Sbjct: 182 VKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFLKPGSHTWTAFAG 361
Query: 1085 SYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYPHEVLLCLESREL-----DQHFID 1139
++GY APELA TM+V EKCDVYSFGV AL+I++G +P +++ L S D ID
Sbjct: 362 TFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPSTVPMVNDLLLID 541
Query: 1140 FLDKRPTPPEGPAIQLLVMVATLILKCVAKDPLSRPTMRQVSQEL-LSSDFWSLSVPFNM 1198
LD+RP E P + ++++A+L L C+ K+P SRPTM QVS+ L L +L F M
Sbjct: 542 ILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQVSKALVLGKPPLALGYQFPM 721
Query: 1199 ITLQNL 1204
+ + L
Sbjct: 722 VRVGQL 739
>TC12613
Length = 488
Score = 180 bits (457), Expect(2) = 5e-49
Identities = 90/93 (96%), Positives = 90/93 (96%)
Frame = +1
Query: 1105 VYSFGVVALQILVGKYPHEVLLCLESRELDQHFIDFLDKRPTPPEGPAIQLLVMVATLIL 1164
VYSFGVV L ILVGKYPHEVLLCLESRELDQHFIDFLDKRPTPPEGPAIQLLVMVATLIL
Sbjct: 1 VYSFGVVHLPILVGKYPHEVLLCLESRELDQHFIDFLDKRPTPPEGPAIQLLVMVATLIL 180
Query: 1165 KCVAKDPLSRPTMRQVSQELLSSDFWSLSVPFN 1197
KCVAKDPLS PTMRQVSQELLSSDFWSLSVPFN
Sbjct: 181 KCVAKDPLS*PTMRQVSQELLSSDFWSLSVPFN 279
Score = 32.7 bits (73), Expect(2) = 5e-49
Identities = 14/14 (100%), Positives = 14/14 (100%)
Frame = +2
Query: 1198 MITLQNLLDNFACR 1211
MITLQNLLDNFACR
Sbjct: 281 MITLQNLLDNFACR 322
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 165 bits (417), Expect = 5e-41
Identities = 128/399 (32%), Positives = 199/399 (49%), Gaps = 22/399 (5%)
Frame = +3
Query: 745 LDLSHNSLSGAIIPQLEKLTSLEVLNLSHNQLSGGIPS-DLNGLISLQSIDISYNKLEGP 803
LDLS ++L G I + ++T+LE LN+SHN G +PS L+ L L S+D+SYN L G
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSL--LISVDLSYNDLMGK 1781
Query: 804 LPSLEAFHNASEEALVGNSGLCSGPDNGNANLSPCG---GEKSNKDNNHKLIIAIVIPVA 860
LP + G + S D N N S G K++ +I I
Sbjct: 1782 LPESIVKLPHLKSLYFGCNEHMSPEDPANMNSSLINTDYGRCKGKESRFGQVIVIGAITC 1961
Query: 861 ALIILLVSLGLFFIFRRYRK-------AGKAKKDKGSN------RKNSFFIWNHR-NRIE 906
+++ ++ G+ F+ R +K AGK K +N K+ FFI +
Sbjct: 1962 GSLLITLAFGVLFVCRYRQKLIPWEGFAGK-KYPMETNIIFSLPSKDDFFIKSVSIQAFT 2138
Query: 907 FEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTS 966
E I ATE + K IG GG GSVY+ L G AVK + F +
Sbjct: 2139 LEYIEVATERY--KTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQG------TREFDN 2294
Query: 967 EIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLE-RVLQKEQEAKILTWDIRLNM 1025
E++ L+ I+H N++ + G+ + +Y + GSL+ R+ + + KIL W RL++
Sbjct: 2295 ELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSI 2474
Query: 1026 IKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLL-KAGANWTT--P 1082
G A L+Y+H S++HRDI +N+LLD K++DFG ++ + G ++ +
Sbjct: 2475 ALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEV 2654
Query: 1083 VGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYP 1121
G+ GY+ PE T +++EK DV+SFGVV L+I+ G+ P
Sbjct: 2655 RGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREP 2771
Score = 56.2 bits (134), Expect = 3e-08
Identities = 56/196 (28%), Positives = 88/196 (44%)
Frame = +3
Query: 236 NLTILDLTENRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDF 295
N+T++ +++ F GP+ N + Q++ I N T ++G + + L QN
Sbjct: 1353 NVTLVKASKSEF-GPL----LNAYEILQVRPWIEETNQT---DVGVIQKMREELLLQNSG 1508
Query: 296 QGPIPSSIGNLTMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLSMASLT 355
+ S G+ +L L G+ G T L D++ ++L G +P S+A +T
Sbjct: 1509 NRALESWSGDPCIL----LPWKGIACDGSNGSSVITKL---DLSSSNLKGLIPSSIAEMT 1667
Query: 356 RMRELGLSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHLYLYENQ 415
+ L +S N G + LSS LIS+ L ND+ GKLP I +L LY N+
Sbjct: 1668 NLETLNISHNSFDGSVPSFPLSSL--LISVDLSYNDLMGKLPESIVKLPHLKSLYFGCNE 1841
Query: 416 FSGPIPKEIGNLSSIN 431
P N S IN
Sbjct: 1842 HMSPEDPANMNSSLIN 1889
Score = 49.7 bits (117), Expect = 3e-06
Identities = 27/85 (31%), Positives = 44/85 (51%)
Frame = +3
Query: 478 LEELKLSENHLSGPLPSSITHLENIKILHLHWNNFSGSIPEDFGPNFLTNVSFANNSFSG 537
+ +L LS ++L G +PSSI + N++ L++ N+F GS+P + L +V + N G
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSLLISVDLSYNDLMG 1778
Query: 538 NLPSGICRGGNLIYLAANLNNFFGP 562
LP I + +L L N P
Sbjct: 1779 KLPESIVKLPHLKSLYFGCNEHMSP 1853
Score = 47.8 bits (112), Expect = 1e-05
Identities = 31/92 (33%), Positives = 49/92 (52%), Gaps = 1/92 (1%)
Frame = +3
Query: 406 LTHLYLYENQFSGPIPKEIGNLSSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLS 465
+T L L + G IP I ++++ L +S+N F+GS+PS LI++ L N L
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPS-FPLSSLLISVDLSYNDLM 1775
Query: 466 GALPPEIGDLENLEELKLSEN-HLSGPLPSSI 496
G LP I L +L+ L N H+S P+++
Sbjct: 1776 GKLPESIVKLPHLKSLYFGCNEHMSPEDPANM 1871
Score = 42.7 bits (99), Expect = 3e-04
Identities = 38/129 (29%), Positives = 60/129 (46%), Gaps = 3/129 (2%)
Frame = +3
Query: 24 QKEAESLITWMNSLNSPLPSSWKLAGNNTSPCKWTSISCDKAG---TVVEIKLPNAGLDG 80
QK E L+ NS N L SW +G+ W I+CD + + ++ L ++ L G
Sbjct: 1470 QKMREELLL-QNSGNRAL-ESW--SGDPCILLPWKGIACDGSNGSSVITKLDLSSSNLKG 1637
Query: 81 TLNRFDFSAFPNLSNFNVSMNNLVGEIPSGIGNATKLKTLDLGSNNLTNPIPPQIGNLLE 140
+ + NL N+S N+ G +PS ++ L ++DL N+L +P I L
Sbjct: 1638 LIPS-SIAEMTNLETLNISHNSFDGSVPS-FPLSSLLISVDLSYNDLMGKLPESIVKLPH 1811
Query: 141 LQVLIFSNN 149
L+ L F N
Sbjct: 1812 LKSLYFGCN 1838
Score = 41.6 bits (96), Expect = 8e-04
Identities = 26/72 (36%), Positives = 39/72 (54%), Gaps = 2/72 (2%)
Frame = +3
Query: 168 LDLGANYLENPDPDQFKGMKSMTELNLSYNSLTDVPPFVSKCPKLVSLDLSLNTITGKIP 227
LDL ++ L+ P M ++ LN+S+NS P L+S+DLS N + GK+P
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSLLISVDLSYNDLMGKLP 1787
Query: 228 --IHLLTNLKNL 237
I L +LK+L
Sbjct: 1788 ESIVKLPHLKSL 1823
Score = 36.6 bits (83), Expect = 0.024
Identities = 29/109 (26%), Positives = 50/109 (45%), Gaps = 10/109 (9%)
Frame = +3
Query: 521 GPNFLTNVSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGN 580
G + +T + ++++ G +PS I NL L + N+F G +P S + LI V L
Sbjct: 1587 GSSVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVP-SFPLSSLLISVDLSY 1763
Query: 581 NLLSGDITNAFGTYPDLNFIDLGHNQ----------LSGSLSSNWGECK 619
N L G + + P L + G N+ S +++++G CK
Sbjct: 1764 NDLMGKLPESIVKLPHLKSLYFGCNEHMSPEDPANMNSSLINTDYGRCK 1910
Score = 35.8 bits (81), Expect = 0.041
Identities = 28/89 (31%), Positives = 41/89 (45%), Gaps = 1/89 (1%)
Frame = +3
Query: 110 GIGNATKLKTLDLGSNNLTNPIPPQIGNLLELQVLIFSNNSLLKQIPS-QLSNLQNLWLL 168
G ++ + LDL S+NL IP I + L+ L S+NS +PS LS+L L +
Sbjct: 1578 GSNGSSVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSL--LISV 1751
Query: 169 DLGANYLENPDPDQFKGMKSMTELNLSYN 197
DL N L P+ + + L N
Sbjct: 1752 DLSYNDLMGKLPESIVKLPHLKSLYFGCN 1838
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 164 bits (416), Expect = 6e-41
Identities = 98/272 (36%), Positives = 142/272 (52%), Gaps = 5/272 (1%)
Frame = +2
Query: 390 NDMTGKLPPQIGSFHNLTHLYLYENQF---SGPIPKEIGNLSSINDLQLSNNHFNGSIPS 446
N+ G +PP+IG NLT L ++ + SG IP E+G L ++ L L N +GS+
Sbjct: 23 NNYQGGIPPEIG---NLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTP 193
Query: 447 TIGQLKKLITLALDSNQLSGALPPEIGDLENLEELKLSENHLSGPLPSSITHLENIKILH 506
+G LK L ++ L +N LSG +P +L+NL L L N L G +P + + +++L
Sbjct: 194 ELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQ 373
Query: 507 LHWNNFSGSIPEDFGPN-FLTNVSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPE 565
L NNF+GSIP+ G N LT V ++N +G LP +C G L L A N FGPIPE
Sbjct: 374 LWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPE 553
Query: 566 SLRNCTGLIRVLLGNNLLSGDITNAFGTYPDLNFIDLGHNQLSGSLSSNWGECKFLSSFS 625
SL C L R+ +G N L+G I P L ++ N LSG + +
Sbjct: 554 SLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQIT 733
Query: 626 ISSNKVHGNIPPELGKL-RLQNLDLSENNLTG 656
+S+NK+ G +P +G +Q L L N +G
Sbjct: 734 LSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 155 bits (393), Expect = 3e-38
Identities = 98/296 (33%), Positives = 147/296 (49%), Gaps = 1/296 (0%)
Frame = +2
Query: 291 HQNDFQGPIPSSIGNLTMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLS 350
+ N++QG IP IGNLT L R GL+ IPA +G L + + N L+GSL
Sbjct: 17 YYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPE 196
Query: 351 MASLTRMRELGLSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHLY 410
+ L ++ + LS+N LSG++ P NLT L
Sbjct: 197 LGHLKSLKSMDLSNNMLSGQV-------------------------PASFAELKNLTLLN 301
Query: 411 LYENQFSGPIPKEIGNLSSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLSGALPP 470
L+ N+ G IP+ +G + ++ LQL N+F GSIP ++G+ KL + L SN+L+G LPP
Sbjct: 302 LFRNRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPP 481
Query: 471 EIGDLENLEELKLSENHLSGPLPSSITHLENIKILHLHWNNFSGSIPED-FGPNFLTNVS 529
+ L+ L N L GP+P S+ E++ + + N +GSIP+ FG LT V
Sbjct: 482 HMCSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVE 661
Query: 530 FANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNLLSG 585
F +N SG P N+ + + N GP+P ++ N T + ++LL N SG
Sbjct: 662 FQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 137 bits (344), Expect = 1e-32
Identities = 101/319 (31%), Positives = 145/319 (44%), Gaps = 1/319 (0%)
Frame = +2
Query: 125 NNLTNPIPPQIGNLLELQVLIFSNNSLLKQIPSQLSNLQNLWLLDLGANYLENPDPDQFK 184
NN IPP+IGNL +L + L +IP++L LQ L L L N L
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSG------- 181
Query: 185 GMKSMTELNLSYNSLTDVPPFVSKCPKLVSLDLSLNTITGKIPIHLLTNLKNLTILDLTE 244
SLT P + L S+DLS N ++G++P LKNLT+L+L
Sbjct: 182 -------------SLT---PELGHLKSLKSMDLSNNMLSGQVPASF-AELKNLTLLNLFR 310
Query: 245 NRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDFQGPIPSSIG 304
NR G IPE + + L+ L+L NN G+IP +G L +++L N G +P +
Sbjct: 311 NRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMC 490
Query: 305 NLTMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLSMASLTRMRELGLSS 364
+ LQ L + L IP +G C +L + M N L GS+P + L ++ ++
Sbjct: 491 SGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQD 670
Query: 365 NQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSF-HNLTHLYLYENQFSGPIPKE 423
N LSGE P+ GS HN+ + L N+ SGP+P
Sbjct: 671 NLLSGEF--------------------------PETGSVSHNIGQITLSNNKLSGPLPST 772
Query: 424 IGNLSSINDLQLSNNHFNG 442
IGN +S+ L L N F+G
Sbjct: 773 IGNFTSMQKLLLDGNKFSG 829
Score = 133 bits (335), Expect = 1e-31
Identities = 89/271 (32%), Positives = 139/271 (50%), Gaps = 1/271 (0%)
Frame = +2
Query: 533 NSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNLLSGDITNAFG 592
N++ G +P I L+ A G IP L L + L N+LSG +T G
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 593 TYPDLNFIDLGHNQLSGSLSSNWGECKFLSSFSISSNKVHGNIPPELGKL-RLQNLDLSE 651
L +DL +N LSG + +++ E K L+ ++ N++HG IP +G++ L+ L L E
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWE 382
Query: 652 NNLTGNIPVELFNSSSKMLKLNLSNNYLSGHMPTRIGELSELQYLDFSANNLSGPIPNAL 711
NN TG+IP L + K+ ++LS+N L+G +P + + LQ L N L GPIP +L
Sbjct: 383 NNFTGSIPQSL-GKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESL 559
Query: 712 GNCGNLIFLKLSMNNLEGPMPHELGNLVNLQPLLDLSHNSLSGAIIPQLEKLTSLEVLNL 771
G C +L +++ N L G +P L L L + + N LSG ++ + L
Sbjct: 560 GKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQV-EFQDNLLSGEFPETGSVSHNIGQITL 736
Query: 772 SHNQLSGGIPSDLNGLISLQSIDISYNKLEG 802
S+N+LSG +PS + S+Q + + NK G
Sbjct: 737 SNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 128 bits (321), Expect = 6e-30
Identities = 89/278 (32%), Positives = 139/278 (49%), Gaps = 1/278 (0%)
Frame = +2
Query: 93 LSNFNVSMNNLVGEIPSGIGNATKLKTLDLGSNNLTNPIPPQIGNLLELQVLIFSNNSLL 152
L F+ + L GEIP+ +G KL TL L N L+ + P++G+L L+ + SNN L
Sbjct: 71 LLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLS 250
Query: 153 KQIPSQLSNLQNLWLLDLGANYLENPDPDQFKGMKSMTELNLSYNSLT-DVPPFVSKCPK 211
Q+P+ + L+NL LL+L N L P+ M ++ L L N+ T +P + K K
Sbjct: 251 GQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGK 430
Query: 212 LVSLDLSLNTITGKIPIHLLTNLKNLTILDLTENRFEGPIPEEIKNLSNLKQLKLGINNL 271
L +DLS N +TG +P H+ + + T++ L F GPIPE + +L ++++G N L
Sbjct: 431 LTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLF-GPIPESLGKCESLTRIRMGQNFL 607
Query: 272 NGTIPDEIGHLSHLEVLELHQNDFQGPIPSSIGNLTMLQRLHLRLSGLNSSIPAGIGFCT 331
NG+IP + L L +E N G P + S+ IG T
Sbjct: 608 NGSIPKGLFGLPKLTQVEFQDNLLSGEFPET------------------GSVSHNIGQIT 733
Query: 332 NLYFVDMAGNSLTGSLPLSMASLTRMRELGLSSNQLSG 369
++ N L+G LP ++ + T M++L L N+ SG
Sbjct: 734 ------LSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 124 bits (312), Expect = 7e-29
Identities = 91/295 (30%), Positives = 134/295 (44%), Gaps = 2/295 (0%)
Frame = +2
Query: 462 NQLSGALPPEIGDLENLEELKLSENHLSGPLPSSITHLENIKILHLHWNNFSGSIPEDFG 521
N G +PPEIG+L L + LSG +P+ + L+ + L L N SGS+ + G
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 522 P-NFLTNVSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGN 580
L ++ +NN SG +P+ NL L N G IPE +
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFV------------- 343
Query: 581 NLLSGDITNAFGTYPDLNFIDLGHNQLSGSLSSNWGECKFLSSFSISSNKVHGNIPPEL- 639
G P L + L N +GS+ + G+ L+ +SSNK+ G +PP +
Sbjct: 344 -----------GEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMC 490
Query: 640 GKLRLQNLDLSENNLTGNIPVELFNSSSKMLKLNLSNNYLSGHMPTRIGELSELQYLDFS 699
RLQ L N L G IP L S + ++ + N+L+G +P + L +L ++F
Sbjct: 491 SGNRLQTLIALGNFLFGPIPESLGKCES-LTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQ 667
Query: 700 ANNLSGPIPNALGNCGNLIFLKLSMNNLEGPMPHELGNLVNLQPLLDLSHNSLSG 754
N LSG P N+ + LS N L GP+P +GN ++Q LL L N SG
Sbjct: 668 DNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLL-LDGNKFSG 829
Score = 120 bits (302), Expect = 1e-27
Identities = 89/274 (32%), Positives = 126/274 (45%), Gaps = 2/274 (0%)
Frame = +2
Query: 557 NNFFGPIPESLRNCTGLIRVLLGNNLLSGDITNAFGTYPDLNFIDLGHNQLSGSLSSNWG 616
NN+ G IP + N T L+R D + LSG + + G
Sbjct: 23 NNYQGGIPPEIGNLTQLLR------------------------FDAAYCGLSGEIPAELG 130
Query: 617 ECKFLSSFSISSNKVHGNIPPELGKLR-LQNLDLSENNLTGNIPVELFNSSSKMLKLNLS 675
+ + L + + N + G++ PELG L+ L+++DLS N L+G +P F + LNL
Sbjct: 131 KLQKLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLSGQVPAS-FAELKNLTLLNLF 307
Query: 676 NNYLSGHMPTRIGELSELQYLDFSANNLSGPIPNALGNCGNLIFLKLSMNNLEGPMPHEL 735
N L G +P +GE+ L+ L NN +G IP +LG G L + LS N L G +P +
Sbjct: 308 RNRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHM 487
Query: 736 GNLVNLQPLLDLSHNSLSGAIIPQLEKLTSLEVLNLSHNQLSGGIPSDLNGLISLQSIDI 795
+ LQ L+ L N L G I L K SL + + N L+G IP L GL L ++
Sbjct: 488 CSGNRLQTLIALG-NFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEF 664
Query: 796 SYNKLEGPLPSL-EAFHNASEEALVGNSGLCSGP 828
N L G P HN + L N SGP
Sbjct: 665 QDNLLSGEFPETGSVSHNIGQITLSNNK--LSGP 760
Score = 39.7 bits (91), Expect = 0.003
Identities = 24/89 (26%), Positives = 41/89 (45%)
Frame = +2
Query: 61 SCDKAGTVVEIKLPNAGLDGTLNRFDFSAFPNLSNFNVSMNNLVGEIPSGIGNATKLKTL 120
S K ++ I++ L+G++ + F P L+ N L GE P + + +
Sbjct: 554 SLGKCESLTRIRMGQNFLNGSIPKGLFG-LPKLTQVEFQDNLLSGEFPETGSVSHNIGQI 730
Query: 121 DLGSNNLTNPIPPQIGNLLELQVLIFSNN 149
L +N L+ P+P IGN +Q L+ N
Sbjct: 731 TLSNNKLSGPLPSTIGNFTSMQKLLLDGN 817
Score = 30.0 bits (66), Expect = 2.3
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = +2
Query: 92 NLSNFNVSMNNLVGEIPSGIGNATKLKTLDLGSNNLT 128
N+ +S N L G +PS IGN T ++ L L N +
Sbjct: 716 NIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFS 826
Score = 29.3 bits (64), Expect = 3.9
Identities = 25/108 (23%), Positives = 50/108 (46%)
Frame = -3
Query: 183 FKGMKSMTELNLSYNSLTDVPPFVSKCPKLVSLDLSLNTITGKIPIHLLTNLKNLTILDL 242
++G+K++ ++ + S + +L L +L +G++ + L +L+ + D
Sbjct: 517 YEGLKTIPGTHVRWKSPCQLVGRKINEGELPILPQTLRNASGEVVLPELQHLQRRHLSDE 338
Query: 243 TENRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLEL 290
NR P+PEE++ + QL NL G +G + E LE+
Sbjct: 337 LRNRSVEPVPEEVQQ-GKILQLSEARRNLAG--EHVVG*IHGFEALEV 203
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 156 bits (395), Expect = 2e-38
Identities = 84/217 (38%), Positives = 125/217 (56%), Gaps = 4/217 (1%)
Frame = +1
Query: 907 FEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTS 966
F ++ AT NF E IG GG G VYK L TG+ AVK+L + G+Q F
Sbjct: 421 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQ----GFQ--EFVM 582
Query: 967 EIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLE-RVLQKEQEAKILTWDIRLNM 1025
E+ L+ + H N++++ G+ +YE++ GSLE + + + + L W R+ +
Sbjct: 583 EVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKV 762
Query: 1026 IKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLLKAGANW---TTP 1082
G A L Y+H P +++RD+ N+LLD+E+ PKLSDFG A+L G N T
Sbjct: 763 AVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRV 942
Query: 1083 VGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGK 1119
+G+YGY APE A++ K+T K D+YSFGVV L++L G+
Sbjct: 943 MGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGR 1053
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 154 bits (388), Expect = 1e-37
Identities = 117/368 (31%), Positives = 173/368 (46%), Gaps = 29/368 (7%)
Frame = +1
Query: 842 KSNKDNNHKLIIAIVIPVAALIILLVSLGLFFIFRRYRKAGKAKKDKGSNRKNSFFIWNH 901
++ K N K + ++ + A +I + LGL RK K + D+G I NH
Sbjct: 1429 RNKKSINTKKLAGSLVVIIAFVIFITILGLAISTCIQRKKNK-RGDEGE-----IGIINH 1590
Query: 902 ------------RNRIEFEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQ 949
+F I +AT +FS +G GG G VYK +L G AVKRL
Sbjct: 1591 WKDKRGDEDIDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSN 1770
Query: 950 NEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQ 1009
G + F +EI + ++HRN++K++G S E+ ++L
Sbjct: 1771 TS------GQGMEEFKNEIKLIARLQHRNLVKLFGCSVHQD------ENSHANKKMKILL 1914
Query: 1010 KEQEAKILTWDIRLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGT 1069
+K++ W+ RL +I G+A L Y+H D I+HRD+ +N+LLD E PK+SDFG
Sbjct: 1915 DSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGL 2094
Query: 1070 ARLL---KAGANWTTPVGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKY------ 1120
AR+ + A +G+YGYM PE A+ + K DV+SFGV+ L+I+ GK
Sbjct: 2095 ARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYD 2274
Query: 1121 PHEVL--------LCLESRELDQHFIDFLDKRPTPPEGPAIQLLVMVATLILKCVAKDPL 1172
PH L L +E R L+ +D L P P I + VA L CV + P
Sbjct: 2275 PHHHLNLLSHAWRLWIEERPLE--LVDELLDDPVIP--TEILRYIHVALL---CVQRRPE 2433
Query: 1173 SRPTMRQV 1180
+RP M +
Sbjct: 2434 NRPDMLSI 2457
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 149 bits (377), Expect = 2e-36
Identities = 86/249 (34%), Positives = 134/249 (53%), Gaps = 12/249 (4%)
Frame = +1
Query: 908 EDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTSE 967
+++ +AT NF+ +G GG GSVY L G AVKRL D F E
Sbjct: 37 KELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADM------EFAVE 198
Query: 968 IHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQKEQEAK-ILTWDIRLNMI 1026
+ L +RH+N++ + G+ +Y+++ SL L + ++ +L W+ R+N+
Sbjct: 199 VEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIA 378
Query: 1027 KGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLLKAGANWTTP--VG 1084
G A + Y+HH TP I+HRDI +NVLLDS+++ +++DFG A+L+ GA T G
Sbjct: 379 IGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKG 558
Query: 1085 SYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYPHEVLLCLESRELD---------Q 1135
+ GY+APE A+ K E CDV+SFG++ L++ GK P E L R ++ +
Sbjct: 559 TLGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAK 738
Query: 1136 HFIDFLDKR 1144
F +F D R
Sbjct: 739 KFTEFADPR 765
>TC14163
Length = 1712
Score = 149 bits (375), Expect = 3e-36
Identities = 112/364 (30%), Positives = 174/364 (47%), Gaps = 32/364 (8%)
Frame = +1
Query: 353 SLTRMRELG----LSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTH 408
SL++++ L L+ +SG +P L P+L + ++ N ++G++P IG+ L
Sbjct: 397 SLSKIKNLDGFYLLNLKNISGP-FPGFLFKLPKLQFIYIENNQLSGRIPENIGNLTRLDV 573
Query: 409 LYLYENQFSGPIPKEIGNLSSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLSGAL 468
L L N+F+G IP +G L+ + LQL NN G+IP+TI +LK L L+L+ NQ SGA+
Sbjct: 574 LSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQFSGAI 753
Query: 469 PPEIGDLENLEELKLSENHLSGPLPSSITHL-ENIKILHLHWNNFSGSIPEDFGP-NFLT 526
P +L L+LS N SG +P+SI+ L ++ L L N SG IP+ G L
Sbjct: 754 PDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKFRALD 933
Query: 527 NVSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNLLSGD 586
+ ++N FSG +P+ + L N P PE N G+ + L NN+ +
Sbjct: 934 TLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPE--MNVKGIESLDLSNNMFHLN 1107
Query: 587 ITNAFGTYPDL--------------------------NFIDLGHNQLSGSLSSNWGECKF 620
+ T + +FIDL N++SGS ++
Sbjct: 1108QIPKWVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYDFIDLSGNEISGSAVGLVNSTEY 1287
Query: 621 LSSFSISSNKVHGNIPPELGKLRLQNLDLSENNLTGNIPVELFNSSSKMLKLNLSNNYLS 680
L F S NK+ + R + LDLS N + G +P S + + KLN+S N+L
Sbjct: 1288LVGFWGSGNKLKFDFERLKFGERFKYLDLSHNLVFGKVP----KSVAGLEKLNVSYNHLC 1455
Query: 681 GHMP 684
G +P
Sbjct: 1456GEIP 1467
Score = 142 bits (358), Expect = 3e-34
Identities = 116/385 (30%), Positives = 178/385 (46%), Gaps = 33/385 (8%)
Frame = +1
Query: 430 INDLQLSNN------HFNGSIPSTIGQLKKLITL-ALDSNQLSGALPPEIGDLENLEELK 482
+ L LS N F+G+I ++ ++K L L+ +SG P + L L+ +
Sbjct: 328 VTSLYLSGNPENPKSFFSGTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIY 507
Query: 483 LSENHLSGPLPSSITHLENIKILHLHWNNFSGSIPEDFGP-NFLTNVSFANNSFSGNLPS 541
+ N LSG +P +I +L + +L L N F+G+IP G LT + NNS +G +P+
Sbjct: 508 IENNQLSGRIPENIGNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPA 687
Query: 542 GICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNLLSGDITNAFGTY-PDLNFI 600
I R NL YL+ N F G IP+ + T L + L N SG I + T P L ++
Sbjct: 688 TIARLKNLTYLSLEGNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYL 867
Query: 601 DLGHNQLSGSLSSNWGECKFLSSFSISSNKVHGNIPPELGKL-RLQNLDLSENNLTGNIP 659
+LGHNQLSG + G+ + L + +SSN+ G +P L ++ NL+L+ N L P
Sbjct: 868 ELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFP 1047
Query: 660 ------VELFNSSSKMLKLN------LSNNYLSGHMPTRIGELSELQ-----------YL 696
+E + S+ M LN S+ + R G +L ++
Sbjct: 1048EMNVKGIESLDLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYDFI 1227
Query: 697 DFSANNLSGPIPNALGNCGNLIFLKLSMNNLEGPMPHELGNLVNLQPLLDLSHNSLSGAI 756
D S N +SG + + L+ S N L+ E LDLSHN + G +
Sbjct: 1228DLSGNEISGSAVGLVNSTEYLVGFWGSGNKLK--FDFERLKFGERFKYLDLSHNLVFGKV 1401
Query: 757 IPQLEKLTSLEVLNLSHNQLSGGIP 781
+ + LE LN+S+N L G IP
Sbjct: 1402---PKSVAGLEKLNVSYNHLCGEIP 1467
Score = 137 bits (345), Expect = 1e-32
Identities = 111/360 (30%), Positives = 173/360 (47%), Gaps = 3/360 (0%)
Frame = +1
Query: 231 LTNLKNLTILDLTENRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLEL 290
L NLKN++ GP P + L L+ + + N L+G IP+ IG+L+ L+VL L
Sbjct: 433 LLNLKNIS----------GPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLTRLDVLSL 582
Query: 291 HQNDFQGPIPSSIGNLTMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLS 350
N F G IPSS+G LT L +L L + L +IPA I NL ++ + GN +G++P
Sbjct: 583 TGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQFSGAIPDF 762
Query: 351 MASLTRMRELGLSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHLY 410
+S T + L LS N+ SG++ S+ + P+L L+L N ++GK+P +G F L L
Sbjct: 763 FSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKFRALDTLD 942
Query: 411 LYENQFSGPIPKEIGNLSSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLS-GALP 469
L N+FSG +P NL+ I +L L+NN P +K + +L L +N +P
Sbjct: 943 LSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPEM--NVKGIESLDLSNNMFHLNQIP 1116
Query: 470 PEIGDLENLEELKLSENHLSGPLPS-SITHLENIKILHLHWNNFSGS-IPEDFGPNFLTN 527
+ + LKL+ + L + L N SGS + +L
Sbjct: 1117KWVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYDFIDLSGNEISGSAVGLVNSTEYLVG 1296
Query: 528 VSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNLLSGDI 587
+ N + + G YL + N FG +P+S+ GL ++ + N L G+I
Sbjct: 1297FWGSGNKLKFDF-ERLKFGERFKYLDLSHNLVFGKVPKSV---AGLEKLNVSYNHLCGEI 1464
Score = 134 bits (336), Expect = 1e-31
Identities = 107/336 (31%), Positives = 149/336 (43%), Gaps = 4/336 (1%)
Frame = +1
Query: 91 PNLSNFNVSMNNLVGEIPSGIGNATKLKTLDLGSNNLTNPIPPQIGNLLELQVLIFSNNS 150
P L + N L G IP IGN T+L L L N T IP +G L L L NNS
Sbjct: 487 PKLQFIYIENNQLSGRIPENIGNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNS 666
Query: 151 LLKQIPSQLSNLQNLWLLDLGANYLENPDPDQFKGMKSMTELNLSYNSLT-DVPPFVSK- 208
L IP+ ++ L+NL L L N PD F + L LS N + +P +S
Sbjct: 667 LTGTIPATIARLKNLTYLSLEGNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTL 846
Query: 209 CPKLVSLDLSLNTITGKIPIHLLTNLKNLTILDLTENRFEGPIPEEIKNLSNLKQLKLGI 268
PKL L+L N ++GKIP L + L LDL+ NRF G +P KNL+ + L L
Sbjct: 847 APKLRYLELGHNQLSGKIP-DFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLAN 1023
Query: 269 NNLNGTIPDEIGHLSHLEVLELHQNDFQ-GPIPSSIGNLTMLQRLHLRLSGLNSSIPAGI 327
N L P+ ++ +E L+L N F IP + + ++ L L G+ +
Sbjct: 1024NLLVDPFPEM--NVKGIESLDLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDWK 1197
Query: 328 GFCTNLY-FVDMAGNSLTGSLPLSMASLTRMRELGLSSNQLSGELYPSLLSSWPELISLQ 386
T Y F+D++GN ++GS + S + S N+L + L L
Sbjct: 1198PAETYFYDFIDLSGNEISGSAVGLVNSTEYLVGFWGSGNKLKFDF--ERLKFGERFKYLD 1371
Query: 387 LQVNDMTGKLPPQIGSFHNLTHLYLYENQFSGPIPK 422
L N + GK+P + L Y N G IPK
Sbjct: 1372LSHNLVFGKVPKSVAGLEKLNVSY---NHLCGEIPK 1470
Score = 122 bits (307), Expect = 3e-28
Identities = 112/407 (27%), Positives = 178/407 (43%), Gaps = 11/407 (2%)
Frame = +1
Query: 52 TSPCKWTSISC---DKAGTVVEI----KLPNAGLDGTLNRFDFSAFPNLSNFNV-SMNNL 103
T C W ++C DK T + + + P + GT++ S NL F + ++ N+
Sbjct: 277 TDCCTWQGVTCLFDDKRVTSLYLSGNPENPKSFFSGTISP-SLSKIKNLDGFYLLNLKNI 453
Query: 104 VGEIPSGIGNATKLKTLDLGSNNLTNPIPPQIGNLLELQVLIFSNNSLLKQIPSQLSNLQ 163
G P + KL+ + + +N L+ IP IGNL L VL + N IPS + L
Sbjct: 454 SGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLTRLDVLSLTGNRFTGTIPSSVGGLT 633
Query: 164 NLWLLDLGANYLENPDPDQFKGMKSMTELNLSYNSLTD-VPPFVSKCPKLVSLDLSLNTI 222
+L L LG N L P +K++T L+L N + +P F S L L LS N
Sbjct: 634 HLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQFSGAIPDFFSSFTDLGILRLSRNKF 813
Query: 223 TGKIPIHLLTNLKNLTILDLTENRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHL 282
+GKIP + T L L+L N+ G IP+ + L L L N +GT+P +L
Sbjct: 814 SGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNL 993
Query: 283 SHLEVLELHQNDFQGPIPSSIGNLTMLQRLHLRLSGLN-SSIPAGIGFCTNLYFVDMAGN 341
+ + L L N P P N+ ++ L L + + + IP + ++ + +A
Sbjct: 994 TKIFNLNLANNLLVDPFPEM--NVKGIESLDLSNNMFHLNQIPKWVTSSPIIFSLKLARC 1167
Query: 342 SLTGSLPLSMASLTRMRE-LGLSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQI 400
+ L + T + + LS N++SG L++S L+ N + ++
Sbjct: 1168GIKMKLDDWKPAETYFYDFIDLSGNEISGSAV-GLVNSTEYLVGFWGSGNKLKFDF-ERL 1341
Query: 401 GSFHNLTHLYLYENQFSGPIPKEIGNLSSINDLQLSNNHFNGSIPST 447
+L L N G +PK + L +N +S NH G IP T
Sbjct: 1342KFGERFKYLDLSHNLVFGKVPKSVAGLEKLN---VSYNHLCGEIPKT 1473
Score = 116 bits (290), Expect = 2e-26
Identities = 82/265 (30%), Positives = 119/265 (43%), Gaps = 7/265 (2%)
Frame = +1
Query: 550 IYLAANLNN----FFGPIPESL---RNCTGLIRVLLGNNLLSGDITNAFGTYPDLNFIDL 602
+YL+ N N F G I SL +N G + L N +SG P L FI +
Sbjct: 337 LYLSGNPENPKSFFSGTISPSLSKIKNLDGFYLLNLKN--ISGPFPGFLFKLPKLQFIYI 510
Query: 603 GHNQLSGSLSSNWGECKFLSSFSISSNKVHGNIPPELGKLRLQNLDLSENNLTGNIPVEL 662
+NQLSG + N G L S++ N+ G IP +G L
Sbjct: 511 ENNQLSGRIPENIGNLTRLDVLSLTGNRFTGTIPSSVGGL-------------------- 630
Query: 663 FNSSSKMLKLNLSNNYLSGHMPTRIGELSELQYLDFSANNLSGPIPNALGNCGNLIFLKL 722
+ + +L L NN L+G +P I L L YL N SG IP+ + +L L+L
Sbjct: 631 ----THLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQFSGAIPDFFSSFTDLGILRL 798
Query: 723 SMNNLEGPMPHELGNLVNLQPLLDLSHNSLSGAIIPQLEKLTSLEVLNLSHNQLSGGIPS 782
S N G +P + L L+L HN LSG I L K +L+ L+LS N+ SG +P+
Sbjct: 799 SRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPA 978
Query: 783 DLNGLISLQSIDISYNKLEGPLPSL 807
L + +++++ N L P P +
Sbjct: 979 SFKNLTKIFNLNLANNLLVDPFPEM 1053
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 147 bits (370), Expect = 1e-35
Identities = 91/251 (36%), Positives = 143/251 (56%), Gaps = 7/251 (2%)
Frame = +2
Query: 882 GKAKKDKGSNRKNSFFIWNHRNRI-EFEDICTATENFSEKYSIGTGGQGSVYKAVLPTGD 940
G K ++G SF N+ RI ++++ AT FS+ +G GG GSVY G
Sbjct: 203 GSEKVEEGPT---SFGSVNNSWRIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGL 373
Query: 941 IFAVKRL---HQNEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFSYLNGSMFFIYE 997
AVK+L + EME F E+ L +RH+N++ + G+ + +Y+
Sbjct: 374 QIAVKKLKAMNSKAEME---------FAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYD 526
Query: 998 HVEKGSLERVLQKEQEAKI-LTWDIRLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLL 1056
++ SL L + ++ L W R+ + G A + Y+HH+ TP I+HRDI +NVLL
Sbjct: 527 YMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLL 706
Query: 1057 DSEYEPKLSDFGTARLLKAG-ANWTTPV-GSYGYMAPELALTMKVTEKCDVYSFGVVALQ 1114
+S++EP ++DFG A+L+ G ++ TT V G+ GY+APE A+ KV+E CDVYSFG++ L+
Sbjct: 707 NSDFEPLVADFGFAKLIPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLE 886
Query: 1115 ILVGKYPHEVL 1125
++ G+ P E L
Sbjct: 887 LVTGRKPIEKL 919
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 142 bits (359), Expect = 2e-34
Identities = 95/297 (31%), Positives = 151/297 (49%), Gaps = 17/297 (5%)
Frame = +1
Query: 907 FEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTS 966
++++ AT NFS IG GG G+VY A L G A+K++ E F
Sbjct: 1057 YQELAKATNNFSLDNKIGQGGFGAVYYAEL-RGKKTAIKKMDVQASTE---------FLC 1206
Query: 967 EIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQKEQEAKILTWDIRLNMI 1026
E+ LT++ H N++++ G+ + GS+F +YEH++ G+L + L + L W R+ +
Sbjct: 1207 ELKVLTHVHHLNLVRLIGYC-VEGSLFLVYEHIDNGNLGQYLHGSGKEP-LPWSSRVQIA 1380
Query: 1027 KGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLLKAGAN--WTTPVG 1084
A L YIH P +HRD+ N+L+D K++DFG +L++ G + T VG
Sbjct: 1381 LDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVG 1560
Query: 1085 SYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYPHEVL----LCLESRELDQHFIDF 1140
++GYM PE A ++ K DVY+FGVV +++ K + VL L ES+ L F +
Sbjct: 1561 TFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAK--NAVLKTGELVAESKGLVALFEEA 1734
Query: 1141 LDKR-----------PTPPEGPAIQLLVMVATLILKCVAKDPLSRPTMRQVSQELLS 1186
L+K P E I ++ +A L C +PL RP+MR + L++
Sbjct: 1735 LNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALMT 1905
>AI967315
Length = 1308
Score = 136 bits (343), Expect = 2e-32
Identities = 79/220 (35%), Positives = 125/220 (55%), Gaps = 5/220 (2%)
Frame = +1
Query: 907 FEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTS 966
+E++ AT FS + +G GG VYK L +GD AVKRL + E + K F +
Sbjct: 118 YEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDER----KEKEFLT 285
Query: 967 EIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQKEQEAKILTWDIRLNMI 1026
EI + ++ H NV+ + G NG ++ ++E GS+ ++ E+ A L W R ++
Sbjct: 286 EIGTIGHVCHSNVMPLLGCCIDNG-LYLVFELSTVGSVASLIHDEKMAP-LDWKTRYKIV 459
Query: 1027 KGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLLKAGANWT----TP 1082
G A L Y+H C I+HRDI +N+LL ++EP++SDFG A+ L + WT P
Sbjct: 460 LGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLP--SQWTHHSIAP 633
Query: 1083 V-GSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYP 1121
+ G++G++APE + V EK DV++FGV L+++ G+ P
Sbjct: 634 IEGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKP 753
>CN825263
Length = 663
Score = 135 bits (339), Expect = 5e-32
Identities = 74/199 (37%), Positives = 113/199 (56%), Gaps = 4/199 (2%)
Frame = +1
Query: 927 GQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFS 986
G G VYK +L G AVK L ++++ + F +E+ L+ + HRN++K+ G
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRG------GREFLAEVEMLSRLHHRNLVKLIGIC 162
Query: 987 YLNGSMFFIYEHVEKGSLERVLQ-KEQEAKILTWDIRLNMIKGLANALSYIHHDCTPSIV 1045
+ IYE V GS+E L ++E L W+ R+ + G A L+Y+H D P ++
Sbjct: 163 IEKQTRCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVI 342
Query: 1046 HRDISGNNVLLDSEYEPKLSDFGTARLLKAGAN---WTTPVGSYGYMAPELALTMKVTEK 1102
HRD +N+LL+ ++ PK+SDFG AR N T +G++GY+APE A+T + K
Sbjct: 343 HRDFKSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVK 522
Query: 1103 CDVYSFGVVALQILVGKYP 1121
DVYS+GVV L++L G P
Sbjct: 523 SDVYSYGVVLLELLTGTKP 579
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 134 bits (337), Expect = 9e-32
Identities = 66/165 (40%), Positives = 104/165 (63%), Gaps = 12/165 (7%)
Frame = +3
Query: 967 EIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQKEQEA---------KIL 1017
E+ L+NIRH N++K+ S+ +Y+++E SL+R L K+ + I+
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 1018 TWDIRLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARLL---K 1074
W RL++ G+A L Y+HHDC+P +VHRD+ +N+LLDS++ K++DFG A + +
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 1075 AGANWTTPVGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGK 1119
A + G++GY+APE ALT++V EK DVYSFGV+ L++ GK
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGK 497
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 132 bits (332), Expect = 3e-31
Identities = 92/301 (30%), Positives = 147/301 (48%), Gaps = 21/301 (6%)
Frame = +2
Query: 905 IEFEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQNEEMEDFPGYQAKNF 964
+ +++ T+NF K IG G G VY A L G+ AVK+L + E E F
Sbjct: 323 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPET-----NNEF 487
Query: 965 TSEIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVL------QKEQEAKILT 1018
+++ ++ +++ N ++++G+ YE GSL +L Q Q L
Sbjct: 488 LTQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLD 667
Query: 1019 WDIRLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARL---LKA 1075
W R+ + A L Y+H P+I+HRDI +NVL+ +Y+ K++DF + + A
Sbjct: 668 WIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAA 847
Query: 1076 GANWTTPVGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYP--------HEVLLC 1127
+ T +G++GY APE A+T ++T+K DVYSFGVV L++L G+ P + L+
Sbjct: 848 RLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVT 1027
Query: 1128 LESRELDQ----HFIDFLDKRPTPPEGPAIQLLVMVATLILKCVAKDPLSRPTMRQVSQE 1183
+ L + +D K PP+G + L VA L CV + RP M V +
Sbjct: 1028 WATPRLSEDKVKQCVDPKLKGEYPPKG--VAKLAAVAAL---CVQYEAEFRPNMSIVVKA 1192
Query: 1184 L 1184
L
Sbjct: 1193 L 1195
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 132 bits (332), Expect = 3e-31
Identities = 94/307 (30%), Positives = 149/307 (47%), Gaps = 11/307 (3%)
Frame = +3
Query: 889 GSNRKNSFFIWNHRNRIEFEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLH 948
G K F N + ED+ A+ +G G G+ YKAVL TG + AVKRL
Sbjct: 207 GVGAKKLVFFGNSARGFDLEDLLRASAEV-----LGKGTFGTAYKAVLETGLVVAVKRLK 371
Query: 949 QNEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVL 1008
E K F +I + + H++++ + + + +Y+++ GSL +L
Sbjct: 372 DVTISE-------KEFKDKIETVGAMDHQSLVPLRAYYFSRDEKLLVYDYMPMGSLSALL 530
Query: 1009 QKEQEA--KILTWDIRLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSD 1066
+ A L W+IR + G A + Y+H P++ H +I +N+LL YE K+SD
Sbjct: 531 HGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQ-GPNVSHGNIKASNILLTKSYEAKVSD 707
Query: 1067 FGTARLLKAGANWTTPVGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGKYPHEVLL 1126
FG A L+ + TP GY APE+ KV++K DVYSFGV+ L++L GK P LL
Sbjct: 708 FGLAHLVGPSS---TPNRVAGYRAPEVTDPRKVSQKADVYSFGVLLLELLTGKAPTHALL 878
Query: 1127 CLESRELDQHFIDFLDKRPTPPEGPAIQLL---------VMVATLILKCVAKDPLSRPTM 1177
E +L + ++ L + E ++LL V + L + C A P RP++
Sbjct: 879 NEEGVDLPR-WVQSLVREEWTSEVFDLELLRYQNVEEEMVQLLQLAVDCAAPYPDRRPSI 1055
Query: 1178 RQVSQEL 1184
+V++ +
Sbjct: 1056 AEVTRSI 1076
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 130 bits (328), Expect = 9e-31
Identities = 91/290 (31%), Positives = 133/290 (45%), Gaps = 12/290 (4%)
Frame = +1
Query: 842 KSNKDNNHKLIIAIVIPVAALIILLVSLGLFFIFRRYRKAGKAKKDKGSNRKNSFFIWNH 901
++ K N K + + A II + LGL + RK + ++D+G W
Sbjct: 1204 RNKKSINIKKLAGSLAGSIAFIICITILGLATVTCIRRKKNE-REDEGGIETRIINHWKD 1380
Query: 902 RNRIE---------FEDICTATENFSEKYSIGTGGQGSVYKAVLPTGDIFAVKRLHQNEE 952
+ E F I + T +FSE +G GG G VYK VL G AVKRL
Sbjct: 1381 KRGDEDIDLATIFDFSTISSTTNHFSESNKLGEGGFGPVYKGVLANGQEIAVKRLSNTS- 1557
Query: 953 MEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHVEKGSLERVLQKEQ 1012
G + F +E+ + ++HRN++K+ G S + M IYE + SL+
Sbjct: 1558 -----GQGMEEFKNEVKLIARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLD------- 1701
Query: 1013 EAKILTWDIRLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLLDSEYEPKLSDFGTARL 1072
Y D I+HRD+ +N+LLDSE PK+SDFG AR+
Sbjct: 1702 ----------------------YFIFDSRLRIIHRDLKTSNILLDSEMNPKISDFGLARI 1815
Query: 1073 L---KAGANWTTPVGSYGYMAPELALTMKVTEKCDVYSFGVVALQILVGK 1119
+ A +G+YGYM+PE A+ + K DV+SFGV+ L+I+ GK
Sbjct: 1816 FTGDQVEAKTKRVMGTYGYMSPEYAVHGSFSVKSDVFSFGVIVLEIISGK 1965
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 130 bits (328), Expect = 9e-31
Identities = 71/228 (31%), Positives = 128/228 (56%), Gaps = 10/228 (4%)
Frame = +3
Query: 883 KAKKDKGSNRK---NSFFIWNHRNRIEFEDICTATENFSEKYSIGTGGQGSVYKAVLPTG 939
K K+++GSN + + + ++ AT NF EK+ +G GG G VY+ +LP
Sbjct: 57 KKKREEGSNSQILGTLKSLPGTPREFNYVELKKATNNFDEKHKLGQGGYGVVYRGMLPKE 236
Query: 940 DIFAVKRLHQNEEMEDFPGYQAKNFTSEIHALTNIRHRNVIKIYGFSYLNGSMFFIYEHV 999
+ ++ ++M+ +F SE+ + +RH++++++ G+ + NG + +Y+++
Sbjct: 237 KLEVAVKMFSRDKMKS-----TDDFLSELIIINRLRHKHLVRLQGWCHKNGVLLLVYDYM 401
Query: 1000 EKGSLERVLQKEQEAKI---LTWDIRLNMIKGLANALSYIHHDCTPSIVHRDISGNNVLL 1056
GSL+ + E+ I L+W +R +I G+A+AL+Y+H++ +VHRD+ +N++L
Sbjct: 402 PNGSLDSHIFCEEGGTITTPLSWPLRYKIISGVASALNYLHNEYDQKVVHRDLKASNIML 581
Query: 1057 DSEYEPKLSDFGTARLLKAGANWTTPV----GSYGYMAPELALTMKVT 1100
DSE+ KL DFG AR L+ T + G+ GY+APE T K T
Sbjct: 582 DSEFNAKLGDFGLARALENEKISYTELEGVHGTMGYIAPECFHTGKAT 725
>TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-cell
leukemia, homeobox 1), partial (5%)
Length = 1304
Score = 130 bits (327), Expect = 1e-30
Identities = 90/265 (33%), Positives = 137/265 (50%), Gaps = 6/265 (2%)
Frame = +1
Query: 259 SNLKQLKLGINNLNGTIPDEIGHLSHLEVLELHQNDFQGPIPSSIGNLTMLQRLHLRLSG 318
+N+ Q+ L +G++ I L+ L L+L +N F GPIPSS+ +L+ L+ L LR +
Sbjct: 229 NNINQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNS 408
Query: 319 LNSSIPAGIGFCTNLYFVDMAGNSLTGSLPLSMASLTRMRELGLSSNQLSGELYPSLLSS 378
+ SIP I +LY +D + NSL+GSLP SM SL +R + LS N+L+G + P L
Sbjct: 409 FSGSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSI-PKLP-- 579
Query: 379 WPELISLQLQVNDMTGKLPPQIGSFHNLTHL---YLYENQFSGPIPKEIGNLSSINDLQL 435
P L+ L ++ N ++G P Q +F L L L EN +G + L S+ + L
Sbjct: 580 -PNLLELAVKANSLSG--PLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDL 750
Query: 436 SNNHFNG---SIPSTIGQLKKLITLALDSNQLSGALPPEIGDLENLEELKLSENHLSGPL 492
SNN F G S P+ G+ L+ + L N++ G P + L L + N L G +
Sbjct: 751 SNNSFTGVQISRPAR-GEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAI 927
Query: 493 PSSITHLENIKILHLHWNNFSGSIP 517
P ++++K L L N G P
Sbjct: 928 PLEYGGIKSMKRLFLDGNFLDGKPP 1002
Score = 126 bits (316), Expect = 2e-29
Identities = 97/318 (30%), Positives = 150/318 (46%), Gaps = 7/318 (2%)
Frame = +1
Query: 374 SLLSSWPELI---SLQLQVNDMTGKLPPQIGSFHNLTHLYLYENQFSGPIPKEIGNLSSI 430
S L+SW I SL + + + G + +N+ + L +SG + I L+ +
Sbjct: 130 SCLASWNFTIDPCSLPRRTHFICGLTCTATTTTNNINQITLDPAGYSGSLTPLISQLTQL 309
Query: 431 NDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQLSGALPPEIGDLENLEELKLSENHLSG 490
L L+ N F+G IPS++ L L TL L SN SG++PP I L++L L S N LSG
Sbjct: 310 VTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFSGSIPPSITTLKSLYSLDFSHNSLSG 489
Query: 491 PLPSSITHLENIKILHLHWNNFSGSIPEDFGPNFLTNVSFANNSFSGNLPSGICRGGNLI 550
LP+S+ L N++ + L +N +GSIP+ LP NL+
Sbjct: 490 SLPNSMNSLINLRRIDLSFNKLTGSIPK--------------------LPP------NLL 591
Query: 551 YLAANLNNFFGPIPESLRNCTGLIRVL-LGNNLLSGDITNAFGTYPDLNFIDLGHNQLSG 609
LA N+ GP+ ++ + V+ L N L+G + F P L +DL +N +G
Sbjct: 592 ELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNSFTG 771
Query: 610 SLSSN--WGECKFLSSFSISSNKVHGNIPPELGKL-RLQNLDLSENNLTGNIPVELFNSS 666
S GE L + ++ N++ G P L +L +L + N+L G IP+E +
Sbjct: 772 VQISRPARGEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAIPLE-YGGI 948
Query: 667 SKMLKLNLSNNYLSGHMP 684
M +L L N+L G P
Sbjct: 949 KSMKRLFLDGNFLDGKPP 1002
Score = 122 bits (305), Expect = 4e-28
Identities = 84/272 (30%), Positives = 129/272 (46%), Gaps = 5/272 (1%)
Frame = +1
Query: 230 LLTNLKNLTILDLTENRFEGPIPEEIKNLSNLKQLKLGINNLNGTIPDEIGHLSHLEVLE 289
L++ L L LDL EN F GPIP + +LSNLK L L N+ +G+IP I L L L+
Sbjct: 286 LISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFSGSIPPSITTLKSLYSLD 465
Query: 290 LHQNDFQGPIPSSIGNLTMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSLPL 349
N G +P+S+ +L L+R +D++ N LTGS+P
Sbjct: 466 FSHNSLSGSLPNSMNSLINLRR------------------------IDLSFNKLTGSIPK 573
Query: 350 SMASLTRMRELGLSSNQLSGELYPSLLSSWPELISLQLQVNDMTGKLPPQIGSFHNLTHL 409
+L EL + +N LSG L + +L ++L N + G + P +L +
Sbjct: 574 LPPNLL---ELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQV 744
Query: 410 YLYENQFSG-----PIPKEIGNLSSINDLQLSNNHFNGSIPSTIGQLKKLITLALDSNQL 464
L N F+G P E NL ++N L N G P+ + +L +L++ N L
Sbjct: 745 DLSNNSFTGVQISRPARGEGSNLVAVN---LGFNRIQGYAPANLAAYPQLSSLSIRHNSL 915
Query: 465 SGALPPEIGDLENLEELKLSENHLSGPLPSSI 496
GA+P E G +++++ L L N L G P+ +
Sbjct: 916 RGAIPLEYGGIKSMKRLFLDGNFLDGKPPAGL 1011
Score = 108 bits (269), Expect = 7e-24
Identities = 93/298 (31%), Positives = 139/298 (46%), Gaps = 5/298 (1%)
Frame = +1
Query: 55 CKWTSISCDKAGTVVEIKLPNAGLDGTLNRFDFSAFPNLSNFNVSMNNLVGEIPSGIGNA 114
C T + + +I L AG G+L S L +++ N+ G IPS + +
Sbjct: 196 CGLTCTATTTTNNINQITLDPAGYSGSLTPL-ISQLTQLVTLDLAENSFSGPIPSSLSSL 372
Query: 115 TKLKTLDLGSNNLTNPIPPQIGNLLELQVLIFSNNSLLKQIPSQLSNLQNLWLLDLGANY 174
+ LKTL L SN+ + IPP I L L L FS+NSL +P+ +++L NL +DL N
Sbjct: 373 SNLKTLTLRSNSFSGSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNK 552
Query: 175 LENPDPDQFKGMKSMTELNLSYNSLTD--VPPFVSKCPKLVSLDLSLNTITGKI-PIHLL 231
L P K ++ EL + NSL+ +L ++LS NT+ G + P L
Sbjct: 553 LTGSIP---KLPPNLLELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFL 723
Query: 232 TNLKNLTILDLTENRFEG-PIPEEIKNL-SNLKQLKLGINNLNGTIPDEIGHLSHLEVLE 289
L +L +DL+ N F G I + SNL + LG N + G P + L L
Sbjct: 724 --LPSLQQVDLSNNSFTGVQISRPARGEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSLS 897
Query: 290 LHQNDFQGPIPSSIGNLTMLQRLHLRLSGLNSSIPAGIGFCTNLYFVDMAGNSLTGSL 347
+ N +G IP G + ++RL L + L+ PAG+ AG ++TGSL
Sbjct: 898 IRHNSLRGAIPLEYGGIKSMKRLFLDGNFLDGKPPAGL---------VAAGAAVTGSL 1044
Score = 105 bits (261), Expect = 6e-23
Identities = 84/266 (31%), Positives = 127/266 (47%), Gaps = 4/266 (1%)
Frame = +1
Query: 523 NFLTNVSFANNSFSGNLPSGICRGGNLIYLAANLNNFFGPIPESLRNCTGLIRVLLGNNL 582
N + ++ +SG+L I + L+ L N+F GPIP SL + + L + L +N
Sbjct: 229 NNINQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNS 408
Query: 583 LSGDITNAFGTYPDLNFIDLGHNQLSGSLSSNWGECKFLSSFSISSNKVHGNIPPELGKL 642
SG I + T L +D HN LSGSL ++ L +S NK+ G+IP KL
Sbjct: 409 FSGSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIP----KL 576
Query: 643 --RLQNLDLSENNLTGNIPVELFNSSSKMLKLNLSNNYLSGHMPTRIGELSELQYLDFSA 700
L L + N+L+G + F +++ + LS N L+G + L LQ +D S
Sbjct: 577 PPNLLELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSN 756
Query: 701 NNLSGPIPN--ALGNCGNLIFLKLSMNNLEGPMPHELGNLVNLQPLLDLSHNSLSGAIIP 758
N+ +G + A G NL+ + L N ++G P L L L + HNSL GAI
Sbjct: 757 NSFTGVQISRPARGEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSL-SIRHNSLRGAIPL 933
Query: 759 QLEKLTSLEVLNLSHNQLSGGIPSDL 784
+ + S++ L L N L G P+ L
Sbjct: 934 EYGGIKSMKRLFLDGNFLDGKPPAGL 1011
Score = 105 bits (261), Expect = 6e-23
Identities = 70/264 (26%), Positives = 123/264 (46%), Gaps = 2/264 (0%)
Frame = +1
Query: 92 NLSNFNVSMNNLVGEIPSGIGNATKLKTLDLGSNNLTNPIPPQIGNLLELQVLIFSNNSL 151
N++ + G + I T+L TLDL N+ + PIP + +L L+ L +NS
Sbjct: 232 NINQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSF 411
Query: 152 LKQIPSQLSNLQNLWLLDLGANYLENPDPDQFKGMKSMTELNLSYNSLTDVPPFVSKCPK 211
IP ++ L++L+ LD N L P+ + ++ ++LS+N LT P + P
Sbjct: 412 SGSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIPKLP--PN 585
Query: 212 LVSLDLSLNTITGKIPIHLLTNLKNLTILDLTENRFEGPIPEEIKNLSNLKQLKLGINNL 271
L+ L + N+++G + L L +++L+EN G + L +L+Q+ L N+
Sbjct: 586 LLELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNSF 765
Query: 272 NGT--IPDEIGHLSHLEVLELHQNDFQGPIPSSIGNLTMLQRLHLRLSGLNSSIPAGIGF 329
G G S+L + L N QG P+++ L L +R + L +IP G
Sbjct: 766 TGVQISRPARGEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAIPLEYGG 945
Query: 330 CTNLYFVDMAGNSLTGSLPLSMAS 353
++ + + GN L G P + +
Sbjct: 946 IKSMKRLFLDGNFLDGKPPAGLVA 1017
Score = 95.9 bits (237), Expect = 3e-20
Identities = 68/172 (39%), Positives = 95/172 (54%), Gaps = 2/172 (1%)
Frame = +1
Query: 633 GNIPPELGKL-RLQNLDLSENNLTGNIPVELFNSSSKMLKLNLSNNYLSGHMPTRIGELS 691
G++ P + +L +L LDL+EN+ +G IP L +S S + L L +N SG +P I L
Sbjct: 271 GSLTPLISQLTQLVTLDLAENSFSGPIPSSL-SSLSNLKTLTLRSNSFSGSIPPSITTLK 447
Query: 692 ELQYLDFSANNLSGPIPNALGNCGNLIFLKLSMNNLEGPMPHELGNLVNLQPLLDLSHNS 751
L LDFS N+LSG +PN++ + NL + LS N L G +P NL+ L + NS
Sbjct: 448 SLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIPKLPPNLLELA----VKANS 615
Query: 752 LSGAI-IPQLEKLTSLEVLNLSHNQLSGGIPSDLNGLISLQSIDISYNKLEG 802
LSG + E L LEV+ LS N L+G + L SLQ +D+S N G
Sbjct: 616 LSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNSFTG 771
Score = 88.2 bits (217), Expect = 7e-18
Identities = 55/152 (36%), Positives = 81/152 (53%)
Frame = +1
Query: 677 NYLSGHMPTRIGELSELQYLDFSANNLSGPIPNALGNCGNLIFLKLSMNNLEGPMPHELG 736
+++ G T + + + SG + + L+ L L+ N+ GP+P L
Sbjct: 187 HFICGLTCTATTTTNNINQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLS 366
Query: 737 NLVNLQPLLDLSHNSLSGAIIPQLEKLTSLEVLNLSHNQLSGGIPSDLNGLISLQSIDIS 796
+L NL+ L L NS SG+I P + L SL L+ SHN LSG +P+ +N LI+L+ ID+S
Sbjct: 367 SLSNLKTLT-LRSNSFSGSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLS 543
Query: 797 YNKLEGPLPSLEAFHNASEEALVGNSGLCSGP 828
+NKL G +P L N E A+ NS SGP
Sbjct: 544 FNKLTGSIPKLPP--NLLELAVKANS--LSGP 627
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,766,716
Number of Sequences: 28460
Number of extensions: 321364
Number of successful extensions: 3401
Number of sequences better than 10.0: 360
Number of HSP's better than 10.0 without gapping: 1977
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2680
length of query: 1211
length of database: 4,897,600
effective HSP length: 101
effective length of query: 1110
effective length of database: 2,023,140
effective search space: 2245685400
effective search space used: 2245685400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0082b.15


