

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.11
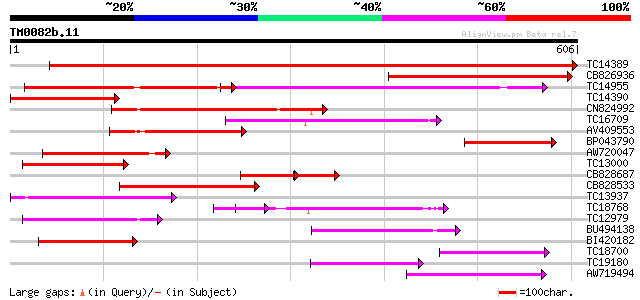
(606 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 1137 0.0
CB826936 317 3e-87
TC14955 UP|O22305 (O22305) Peptide transporter, complete 257 3e-69
TC14390 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 243 8e-65
CN824992 182 1e-46
TC16709 weakly similar to PIR|T47573|T47573 peptide transport-li... 173 6e-44
AV409553 169 9e-43
BP043790 153 7e-38
AW720047 143 7e-35
TC13000 similar to PIR|T47573|T47573 peptide transport-like prot... 140 6e-34
CB828687 131 3e-31
CB828533 127 4e-30
TC13937 weakly similar to UP|Q9LVE0 (Q9LVE0) Nitrate transporter... 119 1e-27
TC18768 119 2e-27
TC12979 similar to UP|Q9LQL2 (Q9LQL2) F5D14.23 protein, partial ... 109 1e-24
BU494138 109 1e-24
BI420182 97 6e-21
TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like p... 95 4e-20
TC19180 similar to PIR|T10255|T10255 nitrite transport protein, ... 91 4e-19
AW719494 82 3e-16
>TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(72%)
Length = 1990
Score = 1137 bits (2941), Expect = 0.0
Identities = 563/564 (99%), Positives = 564/564 (99%)
Frame = +3
Query: 43 IVGYELFERMAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYL 102
+VGYELFERMAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYL
Sbjct: 18 LVGYELFERMAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYL 197
Query: 103 GRYWTFIIASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYI 162
GRYWTFIIASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYI
Sbjct: 198 GRYWTFIIASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYI 377
Query: 163 IALGAGGTKPNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNL 222
IALGAGGTKPNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNL
Sbjct: 378 IALGAGGTKPNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNL 557
Query: 223 SWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPK 282
SWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPK
Sbjct: 558 SWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPK 737
Query: 283 QLHELSMEEYANNSRNRIDHTSTLRFLDKAAVKTGKTSPWRLCTVTQVEETKQMTKMVPI 342
QLHELSMEEYANNSRNRIDHTSTLRFLDKAAVKTGKTSPWRLCTVTQVEETKQMTKMVPI
Sbjct: 738 QLHELSMEEYANNSRNRIDHTSTLRFLDKAAVKTGKTSPWRLCTVTQVEETKQMTKMVPI 917
Query: 343 LITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFV 402
LITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFV
Sbjct: 918 LITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFV 1097
Query: 403 PVIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSI 462
PVIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSI
Sbjct: 1098PVIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSI 1277
Query: 463 FILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVA 522
FILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVA
Sbjct: 1278FILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVA 1457
Query: 523 DITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVTHEKSGSE 582
DITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVTHEKSGSE
Sbjct: 1458DITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVTHEKSGSE 1637
Query: 583 LNPASLDNARICQGSESTAQPDAK 606
LNPASLDNARICQGSESTAQPDAK
Sbjct: 1638LNPASLDNARICQGSESTAQPDAK 1709
>CB826936
Length = 596
Score = 317 bits (813), Expect = 3e-87
Identities = 154/197 (78%), Positives = 173/197 (87%)
Frame = +3
Query: 405 IRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFI 464
+R+YTKNPRGI MLQRLGAGL+MY++ MV AWL+ERKRL VARE +LLGQHD IPL+IFI
Sbjct: 6 MRQYTKNPRGIAMLQRLGAGLVMYIITMVTAWLSERKRLSVAREYNLLGQHDKIPLTIFI 185
Query: 465 LIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADI 524
L+PQ+ALTGVA NF +IA +DFFY QAPEGMKSLGISYSTTS ALGCF SSFLLSTVAD+
Sbjct: 186 LLPQFALTGVANNFVDIAMLDFFYDQAPEGMKSLGISYSTTSTALGCFFSSFLLSTVADL 365
Query: 525 TKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVTHEKSGSELN 584
TKKH H+GW+LDNLN+S LDYYYAFM ILS NFLCFLV AK F YNVDV H+KSG E+N
Sbjct: 366 TKKHGHKGWVLDNLNVSHLDYYYAFMAILSVANFLCFLVAAKLFAYNVDVIHKKSGLEMN 545
Query: 585 PASLDNARICQGSESTA 601
ASLD+ARICQ SESTA
Sbjct: 546 HASLDSARICQSSESTA 596
>TC14955 UP|O22305 (O22305) Peptide transporter, complete
Length = 2300
Score = 257 bits (657), Expect = 3e-69
Identities = 134/352 (38%), Positives = 206/352 (58%), Gaps = 2/352 (0%)
Frame = +1
Query: 226 LGYGLPTVGLAFSILVFLVGTPFYRHKLPSG-SPLTRMLQVFVAAGIKWKAHVPHDPKQL 284
L Y L +G S ++F G+P YRHK SP ++V + A K +P +P +L
Sbjct: 667 LAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSEL 846
Query: 285 HELSMEEYANNSRNRIDHTSTLRFLDKAAVKTGKTSPWRL-CTVTQVEETKQMTKMVPIL 343
HE + Y ++ +I HTS FLD+AA++ T CTVTQVE TK + M I
Sbjct: 847 HEFQLNYYISSGARKIHHTSHFSFLDRAAIRESNTDLSNPPCTVTQVEGTKLVLGMFQIW 1026
Query: 344 ITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVP 403
+ LIP T+F++QGTT+DR++GP F +P A L ++++ L +PIYD F+P
Sbjct: 1027 LLMLIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPIYDHYFIP 1206
Query: 404 VIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIF 463
+RR T N RGI +LQR+G G+ + V+ M + + E +R+ V ++ H++G + +P+SIF
Sbjct: 1207 FMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMSIF 1386
Query: 464 ILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVAD 523
L+PQ + GV+ F ++FFY Q+PE MK LG + T+ VA G ++++FL++ +
Sbjct: 1387 WLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMIDK 1566
Query: 524 ITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVT 575
+ WI +NLN S LDYYYAF+ ++S LNF FL + + Y + T
Sbjct: 1567 L-------NWIGNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENT 1701
Score = 215 bits (547), Expect = 2e-56
Identities = 107/227 (47%), Positives = 148/227 (65%)
Frame = +3
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 75
YT DGTVDL GRPVL S TG KACT+I+ Y + ER AYYG+ +NLV F+ ++L++ V+
Sbjct: 51 YTLDGTVDLAGRPVLSSLTGKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVS 230
Query: 76 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPP 135
S + +NW+G+ TP++GAYIAD Y GR+WT + IY +G+ LL LT +L +LRP
Sbjct: 231 SITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRP-- 404
Query: 136 CAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKLS 195
EN C +AS +FY +LY IA+G+G KPN+ST GADQFD++ +E K+S
Sbjct: 405 ---ACENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVS 575
Query: 196 FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVF 242
FFNWW F+ G L +T +VYIQ+ G+GL + FS ++
Sbjct: 576 FFNWWAFNGACGSLMATLFVVYIQEKN--GXGFGLQFICYWFSTFLY 710
>TC14390 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(18%)
Length = 622
Score = 243 bits (619), Expect = 8e-65
Identities = 117/117 (100%), Positives = 117/117 (100%)
Frame = +1
Query: 1 MSVVDEKGLPSGKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSN 60
MSVVDEKGLPSGKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSN
Sbjct: 37 MSVVDEKGLPSGKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSN 216
Query: 61 LVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLL 117
LVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLL
Sbjct: 217 LVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLL 387
>CN824992
Length = 708
Score = 182 bits (462), Expect = 1e-46
Identities = 101/238 (42%), Positives = 146/238 (60%), Gaps = 8/238 (3%)
Frame = +2
Query: 110 IASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGG 169
I I+++G+ L+LT L L P C G + C S + +FY+++Y+IALG GG
Sbjct: 5 IFQVIFVVGLASLSLTTYLFLLEPNGC--GDKELPCTSHSSYQTALFYVSIYLIALGNGG 178
Query: 170 TKPNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYG 229
+PNI+T GADQFDE +PKE K+SFF+++ ++ +G LFS T+L Y +D+ W+LG+
Sbjct: 179 YQPNIATFGADQFDEEDPKEQHSKISFFSYFYLALNLGSLFSNTILNYFEDDGLWTLGFW 358
Query: 230 LPTVGLAFSILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHV-PHDPKQLHELS 288
++++FL GTP+YR+ P G+P+ R QVFVAA KWK V P + QLHE
Sbjct: 359 ASAGSAFLALVLFLCGTPWYRYFKPGGNPVPRFCQVFVAAMRKWKVKVSPGEEDQLHE-- 532
Query: 289 MEEYANNSRNRIDHTSTLRFLDKAAVKTGKTSP-------WRLCTVTQVEETKQMTKM 339
+EE + N ++ HT RFLDKAA+ T + S W L TVTQVEE K + ++
Sbjct: 533 VEECSTNGGRKMFHTEGFRFLDKAALITSQESKQENECSLWHLSTVTQVEEVKCILRL 706
>TC16709 weakly similar to PIR|T47573|T47573 peptide transport-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(41%)
Length = 712
Score = 173 bits (439), Expect = 6e-44
Identities = 96/238 (40%), Positives = 142/238 (59%), Gaps = 7/238 (2%)
Frame = +1
Query: 231 PTVGLAFSILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSME 290
P V +A +++ F GT YR++ P GSPLTRM QV VA+ K P D L+E++
Sbjct: 1 PAVAMAVAVVSFFSGTRLYRNQKPGGSPLTRMCQVVVASMRKCGVQAPDDKSLLYEIADT 180
Query: 291 EYANNSRNRIDHTSTLRFLDKAAV------KTGKTSPWRLCTVTQVEETKQMTKMVPILI 344
E A ++DHT+ L F DKAAV + PWRLCT TQVEE K + +M+P+
Sbjct: 181 ESAIKGSRKLDHTNELSFFDKAAVVVQSSDQKDSVDPWRLCTETQVEELKSILRMLPVWA 360
Query: 345 TTLIPCTMLIQAHTLFIKQGTTLDRSMG-PNFEIPPAGLSAILIISMLTSIPIYDRVFVP 403
T +I T+ Q TLF+ QG T++ +G NF+IPPA LS +S++ +P+YD + VP
Sbjct: 361 TGIIFATVYGQMSTLFVLQGQTMNTHVGNSNFKIPPAALSIFDTLSVIFWVPVYDWIIVP 540
Query: 404 VIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLS 461
+ R+++ + G+T LQR+G GL + + MV A + E RLR+ R +H + +P+S
Sbjct: 541 IARKFSGHKNGLTQLQRMGIGLFISIFAMVAAAILEVIRLRMVR-RHNYYELKEVPMS 711
>AV409553
Length = 433
Score = 169 bits (429), Expect = 9e-43
Identities = 81/147 (55%), Positives = 104/147 (70%)
Frame = +2
Query: 107 TFIIASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALG 166
TF ++S +Y++GM LLT+ VSL +L+P C G+ C +AS FY ALY A+G
Sbjct: 2 TFTLSSIVYVMGMILLTVAVSLKSLKPT-CTNGI----CNKASTSQIVFFYTALYTTAIG 166
Query: 167 AGGTKPNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSL 226
AGGTKPNIST GADQFD++ P E K SFFNWW+F+ +G L +T LVYIQ+NL W L
Sbjct: 167 AGGTKPNISTFGADQFDDFNPHEKQTKASFFNWWMFTSFLGALIATLGLVYIQENLGWGL 346
Query: 227 GYGLPTVGLAFSILVFLVGTPFYRHKL 253
GYG+PT GL S+++F VGTP YRHK+
Sbjct: 347 GYGIPTSGLLLSLVIFYVGTPMYRHKV 427
>BP043790
Length = 477
Score = 153 bits (387), Expect = 7e-38
Identities = 73/98 (74%), Positives = 82/98 (83%)
Frame = -1
Query: 487 FYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYY 546
FY QAPEGMKSLG SY TTS+ LG FLS+FLLSTVADITK++ H+GW+LDNLNISRLDYY
Sbjct: 477 FYDQAPEGMKSLGTSYFTTSLGLGSFLSTFLLSTVADITKRNGHKGWVLDNLNISRLDYY 298
Query: 547 YAFMVILSFLNFLCFLVTAKFFDYNVDVTHEKSGSELN 584
YAFM +LS LNFLCFLV AK F YNVDV H G E++
Sbjct: 297 YAFMAVLSLLNFLCFLVVAKMFVYNVDVRHSNPGLEMS 184
>AW720047
Length = 419
Score = 143 bits (361), Expect = 7e-35
Identities = 66/137 (48%), Positives = 95/137 (69%)
Frame = +3
Query: 36 SWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGA 95
+WKAC FI+G ER+AYYGI++NLV++L +KL+EGTV+++ NV+ + G + TPL+G+
Sbjct: 3 TWKACPFILGSFFCERLAYYGIATNLVIYLTTKLNEGTVSAAKNVNTFHGTCYLTPLIGS 182
Query: 96 YIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGI 155
++ADAY GRYWT + +YL+GMC+L L+ ++PAL+P C V Q+
Sbjct: 183 FLADAYWGRYWTITVFYAVYLIGMCILILSATIPALKPAECVNSVCTATLAQSV-----X 347
Query: 156 FYLALYIIALGAGGTKP 172
F+L LY IALG GG KP
Sbjct: 348 FFLGLYSIALGTGGIKP 398
>TC13000 similar to PIR|T47573|T47573 peptide transport-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(21%)
Length = 446
Score = 140 bits (353), Expect = 6e-34
Identities = 61/114 (53%), Positives = 87/114 (75%)
Frame = +3
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
++YT+DGTVD G P + +TG+WKAC FI+G E ER+AYYG+S+NLV++ +L++ +
Sbjct: 105 DNYTKDGTVDYLGNPANKKQTGTWKACPFILGNECCERLAYYGMSTNLVLYFKKQLNQHS 284
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVS 127
+S NVSNW+G + TPL+GA++AD+YLGRYWT S IY++GM LLTL+ S
Sbjct: 285 ATASKNVSNWSGTCYITPLIGAFLADSYLGRYWTIACFSIIYVIGMTLLTLSAS 446
>CB828687
Length = 554
Score = 131 bits (330), Expect = 3e-31
Identities = 61/63 (96%), Positives = 62/63 (97%)
Frame = +3
Query: 247 PFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTL 306
PFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTL
Sbjct: 3 PFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTL 182
Query: 307 RFL 309
R +
Sbjct: 183 RLV 191
Score = 97.8 bits (242), Expect = 4e-21
Identities = 47/49 (95%), Positives = 47/49 (95%)
Frame = +1
Query: 304 STLRFLDKAAVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTM 352
S RFLDKAAVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTM
Sbjct: 406 SNCRFLDKAAVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTM 552
>CB828533
Length = 557
Score = 127 bits (320), Expect = 4e-30
Identities = 63/151 (41%), Positives = 94/151 (61%), Gaps = 1/151 (0%)
Frame = +1
Query: 118 GMCLLTLTVSLPALRPPPCAQGV-ENQDCPQASPWTRGIFYLALYIIALGAGGTKPNIST 176
GMCLLT ++P+++ PPC+ ++ +C +AS + +ALY IA+GAGG K ++S
Sbjct: 103 GMCLLTGATTIPSMKAPPCSSVQRQHHECIEASGKQVALLLVALYTIAVGAGGIKSSVSG 282
Query: 177 MGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLA 236
G+DQFD +P+E + FFN + F + G LFS VLVY+QDN+ G+G+P +
Sbjct: 283 FGSDQFDTTDPREEKKMVFFFNRFYFFVSTGSLFSVLVLVYVQDNIGRGWGFGIPAGIML 462
Query: 237 FSILVFLVGTPFYRHKLPSGSPLTRMLQVFV 267
S+ L G P YR+K P GSPLT + +V +
Sbjct: 463 VSLAFLLYGKPLYRYKRPQGSPLTLIWKVLI 555
Score = 30.0 bits (66), Expect = 1.1
Identities = 16/34 (47%), Positives = 18/34 (52%)
Frame = +3
Query: 12 GKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVG 45
G ED VD GRP +SKTG W A I+G
Sbjct: 9 GAEDIA--AAVDYLGRPPDKSKTGGWTAAWLILG 104
>TC13937 weakly similar to UP|Q9LVE0 (Q9LVE0) Nitrate transporter
(AT3g21670/MIL23_23), partial (27%)
Length = 558
Score = 119 bits (298), Expect = 1e-27
Identities = 69/179 (38%), Positives = 101/179 (55%), Gaps = 1/179 (0%)
Frame = +2
Query: 1 MSVVDEKGLPSGKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSN 60
M V G G ED VD GRP +SKTG W A I+G EL +++ GIS N
Sbjct: 26 MVQVASHGEKKGAEDIA--AAVDYLGRPPDKSKTGGWTAAWLILGVELAQKIMVLGISMN 199
Query: 61 LVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMC 120
+V +L L+ T S+ V+N G + L+G ++AD LGRY T I++ I +GMC
Sbjct: 200 IVTYLVGVLNLPTAYSATIVTNCLGTMNLVSLLGGFLADVKLGRYLTIAISATISAVGMC 379
Query: 121 LLTLTVSLPALRPPPCAQ-GVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMG 178
LLT ++P+++ PPC+ ++ +C +AS + +ALY IA+GAGG K ++S G
Sbjct: 380 LLTGATTIPSMKAPPCSSVQRQHHECIEASGKQVALLLVALYTIAVGAGGIKSSVSGFG 556
>TC18768
Length = 738
Score = 119 bits (297), Expect = 2e-27
Identities = 84/236 (35%), Positives = 123/236 (51%), Gaps = 8/236 (3%)
Frame = +2
Query: 242 FLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRID 301
F+ GT HK S L + L + K K P +P+ LH +N +
Sbjct: 92 FITGTS--PHKAAS*PLLLKFLWLHF---FKRKHIYPSNPQMLH---------GDQNNVG 229
Query: 302 --HTSTLRFLDKAAVKTG------KTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTML 353
HT RFLDKA ++ K S WRLC+V QVE+ K + ++PI T++ T+L
Sbjct: 230 QVHTDKFRFLDKACIRVEEAGSNTKKSSWRLCSVGQVEQVKILLSVIPIFSCTIVFNTIL 409
Query: 354 IQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPR 413
Q T ++QG+ +D + +F IPPA L +I I ++ +P+YD FVP RR T +
Sbjct: 410 AQLQTFSVQQGSAMDTHLTKSFHIPPASLQSIPYILLIIVVPLYDTFFVPFARRITGHES 589
Query: 414 GITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQY 469
GI+ L+R+G GL + MV A L E+KR R++ L H+ LSIF + PQ+
Sbjct: 590 GISPLRRIGFGLFLATFSMVSAALLEKKR----RDEAL--NHN-KTLSIFWITPQF 736
Score = 42.0 bits (97), Expect = 3e-04
Identities = 22/60 (36%), Positives = 35/60 (57%)
Frame = +1
Query: 218 IQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHV 277
IQ + +G+G+ +A ++ + GT +YR+K P GS LT + QV VAA KA++
Sbjct: 4 IQTHSGMDVGFGISAAVMAMGLISLISGTFYYRNKPPQGSILTPIAQVLVAAFF*KKAYL 183
>TC12979 similar to UP|Q9LQL2 (Q9LQL2) F5D14.23 protein, partial (25%)
Length = 601
Score = 109 bits (273), Expect = 1e-24
Identities = 60/150 (40%), Positives = 88/150 (58%)
Frame = +3
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
E+ T DG+VD GRP +R+K+G W A T I+ + +A++G+ NLV+FL L +
Sbjct: 156 EEVTLDGSVDWHGRPAIRAKSGRWVAGTIILLNQGLATLAFFGVGVNLVLFLTRVLGQDN 335
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRP 133
++NNVS W G V+ LVGA+++D+Y GRY T I I++LG+ L+L+ L LRP
Sbjct: 336 ADAANNVSKWTGTVYLFSLVGAFLSDSYWGRYKTCAIFQGIFVLGLVFLSLSSYLSLLRP 515
Query: 134 PPCAQGVENQDCPQASPWTRGIFYLALYII 163
C G E C + S G+F Y+I
Sbjct: 516 KGC--GSELLHCGKHSSLEMGMFSYQSYLI 599
>BU494138
Length = 518
Score = 109 bits (272), Expect = 1e-24
Identities = 62/160 (38%), Positives = 91/160 (56%), Gaps = 1/160 (0%)
Frame = +2
Query: 323 RLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGL 382
R+ ++ QVEE K + ++ PI ++ T + Q T + Q +DR +G F+IP L
Sbjct: 11 RVVSIQQVEEIKCLARIFPIWAAGILGFTAMAQQGTFTVSQAMKMDRHIGSKFQIPAGSL 190
Query: 383 SAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKR 442
I I++ +P YDR FVP +RR TK+ GIT+LQR+G G++ VL M++A L E+ R
Sbjct: 191 GVISFITIGLWVPFYDRFFVPAVRRITKHEGGITLLQRIGIGMVFSVLSMIVAGLVEKVR 370
Query: 443 LRVAREK-HLLGQHDIIPLSIFILIPQYALTGVAENFAEI 481
VA + LG I P+S+ L PQ L G+ E F I
Sbjct: 371 RGVANSNPNPLG---IAPMSVMWLAPQLVLMGLCEAFNAI 481
>BI420182
Length = 506
Score = 97.4 bits (241), Expect = 6e-21
Identities = 43/106 (40%), Positives = 68/106 (63%)
Frame = +3
Query: 31 RSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTT 90
R K G FI E+ E++A G S+N++ +L ++LH ++N ++N+ G T
Sbjct: 138 RRKQGGLVTMPFIFANEICEKLAVVGFSTNMISYLTTQLHMPLTKAANTLTNFGGTASLT 317
Query: 91 PLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPPC 136
PL+GA+I+D+Y G++WT +AS +Y +GM LT++ LP LRPPPC
Sbjct: 318 PLLGAFISDSYAGKFWTITMASVLYQIGMVSLTISAVLPQLRPPPC 455
>TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like protein,
partial (18%)
Length = 615
Score = 94.7 bits (234), Expect = 4e-20
Identities = 42/118 (35%), Positives = 68/118 (57%)
Frame = +1
Query: 460 LSIFILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLS 519
LS + L+ QY L GVAE F + ++F Y +AP+ MKS+G +Y+ + LGCF ++ + S
Sbjct: 46 LSAYWLLIQYCLIGVAEVFCIVGLLEFLYEEAPDAMKSIGSAYAALAGGLGCFAATIINS 225
Query: 520 TVADITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVTHE 577
+ +T K + W+ N+N R DY+Y + LS +NF F+ +A + Y +E
Sbjct: 226 IIKSLTGKEGKESWLAQNINTGRFDYFYWILTALSLVNFCIFIYSAHRYKYRTQQVYE 399
>TC19180 similar to PIR|T10255|T10255 nitrite transport protein, chloroplast
- cucumber {Cucumis sativus;} , partial (12%)
Length = 408
Score = 91.3 bits (225), Expect = 4e-19
Identities = 47/121 (38%), Positives = 71/121 (57%)
Frame = +1
Query: 322 WRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAG 381
WRL TV +VEE K + +M PI + ++ T Q T ++Q T+DR + +F+IP
Sbjct: 40 WRLNTVHRVEELKSIIRMGPIWASGILLITAYAQQGTFSLQQAKTMDRHITKSFQIPAGS 219
Query: 382 LSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERK 441
+S II+MLT+ +YDRV + V RR+T RGI+ L R+G G ++ + +A E K
Sbjct: 220 MSVFTIITMLTTTALYDRVLIRVARRFTGLDRGISFLHRMGIGFVISTIATFVAGFVEMK 399
Query: 442 R 442
R
Sbjct: 400 R 402
>AW719494
Length = 473
Score = 81.6 bits (200), Expect = 3e-16
Identities = 45/149 (30%), Positives = 75/149 (50%)
Frame = +3
Query: 425 LLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKM 484
L ++ ++ A L +R R ++ H ++ +S L PQ L G+AE F I +
Sbjct: 6 LFSFLHLVTSATLETIRRKRAIEAGYINNAHGVLNMSAMWLAPQLCLGGIAEAFNAIGQN 185
Query: 485 DFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLD 544
+F+Y + P M + S +A G +SSF+ S V +IT +GW+ DN+N R D
Sbjct: 186 EFYYTEFPRTMSXVASSLLGLGMAAGNIVSSFVFSAVENITSSGGKEGWVSDNINKGRFD 365
Query: 545 YYYAFMVILSFLNFLCFLVTAKFFDYNVD 573
YY + +S LN + +L+ + + VD
Sbjct: 366 KYYWVVXGISGLNLVYYLICSWAYGPTVD 452
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,303,705
Number of Sequences: 28460
Number of extensions: 167923
Number of successful extensions: 1001
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 972
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 978
length of query: 606
length of database: 4,897,600
effective HSP length: 96
effective length of query: 510
effective length of database: 2,165,440
effective search space: 1104374400
effective search space used: 1104374400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0082b.11


