

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
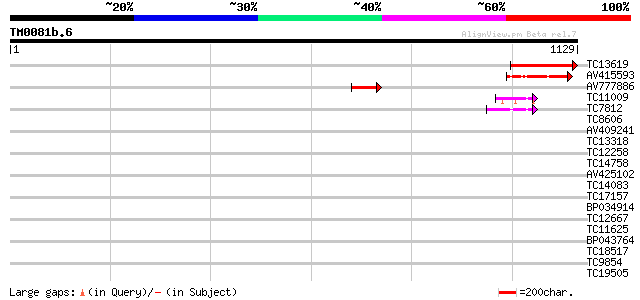
Query= TM0081b.6
(1129 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13619 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulatory... 258 3e-69
AV415593 143 1e-34
AV777886 71 1e-12
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 44 1e-04
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 44 2e-04
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 41 0.001
AV409241 37 0.013
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 37 0.017
TC12258 weakly similar to UP|O50458 (O50458) PGRS_FAMILY protein... 37 0.023
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 35 0.066
AV425102 35 0.066
TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor... 35 0.066
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 35 0.066
BP034914 35 0.087
TC12667 similar to PIR|D96525|D96525 protein T1N15.19 [imported]... 35 0.087
TC11625 weakly similar to UP|O81015 (O81015) At2g26920 protein, ... 35 0.087
BP043764 35 0.087
TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial ... 34 0.11
TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1 (AT3g1... 34 0.11
TC19505 weakly similar to UP|Q9FG35 (Q9FG35) Emb|CAB82953.1, par... 34 0.11
>TC13619 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulatory-like
protein, partial (6%)
Length = 537
Score = 258 bits (660), Expect = 3e-69
Identities = 128/132 (96%), Positives = 129/132 (96%)
Frame = +2
Query: 998 AGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSSLPLSER 1057
AGEDVSGSESAGDECFQE+HEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSSLP SER
Sbjct: 20 AGEDVSGSESAGDECFQEEHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSSLP*SER 199
Query: 1058 FLSSVKPLTKHVSAVSFAEEMKDSRVFYGNDDFYALFRLHQILYERILSAKINSMSAEMK 1117
FLSSVKPLTKHVSAVSFAEEMKDSRV GNDDFYALFRLHQILYERILSAKINSMSAEMK
Sbjct: 200 FLSSVKPLTKHVSAVSFAEEMKDSRVLNGNDDFYALFRLHQILYERILSAKINSMSAEMK 379
Query: 1118 WKAKDASSPDPY 1129
WKAKDASSPDPY
Sbjct: 380 WKAKDASSPDPY 415
>AV415593
Length = 416
Score = 143 bits (361), Expect = 1e-34
Identities = 85/133 (63%), Positives = 99/133 (73%), Gaps = 2/133 (1%)
Frame = +3
Query: 990 EDSENVSEAGEDVSGSESA-GDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG-GG 1047
+DSEN SE DVSGSESA G+EC +E+HE D EHD+ KAESEGEAEGM DA G
Sbjct: 24 DDSENASE-NVDVSGSESADGEECSREEHE--DREHDN---KAESEGEAEGMVDAHDVEG 185
Query: 1048 DSSSLPLSERFLSSVKPLTKHVSAVSFAEEMKDSRVFYGNDDFYALFRLHQILYERILSA 1107
D SL +SERFL +VKPL K+V A + E+ ++S+VFYG+D FY LFRLHQ LYERI SA
Sbjct: 186 DGMSLTISERFLQTVKPLAKYVPA-ALHEKDRNSQVFYGSDSFYVLFRLHQTLYERIQSA 362
Query: 1108 KINSMSAEMKWKA 1120
K NS SAE KWKA
Sbjct: 363 KFNSSSAEKKWKA 401
>AV777886
Length = 540
Score = 70.9 bits (172), Expect = 1e-12
Identities = 33/60 (55%), Positives = 47/60 (78%)
Frame = +3
Query: 680 EFEYSDGGIHEDLYKLVQYSCEEVFSSKELLNKIMRLWSTFLEPMLGVTSQSHGTERVED 739
EF Y D IHEDLY+LV+YSC E+ ++++L +K+M++W+TFLEPML V S++ G E ED
Sbjct: 3 EFHYPDLDIHEDLYQLVKYSCGELCTAEQL-DKVMKVWTTFLEPMLCVPSRAQGAEDTED 179
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 44.3 bits (103), Expect = 1e-04
Identities = 35/103 (33%), Positives = 46/103 (43%), Gaps = 18/103 (17%)
Frame = +2
Query: 967 ESRDREEECGPETG----GDNDADADDEDSENVSEAGEDVSGSE--------SAGDECFQ 1014
+ D EEE G E G GD D D DD+D ++ E E+ G+E +A DE
Sbjct: 113 DEEDEEEEGGVEGGRGGGGDPDDDDDDDDDDDEEEEEEEDLGTEYLVRRTVAAAEDEEAS 292
Query: 1015 EDHEEDDIEHDDVDGKAESEGEAEGM------CDAQGGGDSSS 1051
D E ++ E DD D +GE G+ D G GD S
Sbjct: 293 SDFEPEEDEGDDND---NDDGEKAGVPSKRKRSDKDGSGDDDS 412
Score = 38.1 bits (87), Expect = 0.008
Identities = 18/56 (32%), Positives = 28/56 (49%)
Frame = +2
Query: 970 DREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHD 1025
D +++ GGD+D D +DE+ E E G G D+ +D EE++ E D
Sbjct: 65 DEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDDDDDEEEEEEED 232
Score = 34.3 bits (77), Expect = 0.11
Identities = 19/71 (26%), Positives = 32/71 (44%), Gaps = 7/71 (9%)
Frame = +2
Query: 963 RRKYESRDREEECGPETGGDNDADAD-------DEDSENVSEAGEDVSGSESAGDECFQE 1015
R + + +++ G + GD D D D D+D E+ E G G GD +
Sbjct: 8 RTRMSGDEDDDDDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDD 187
Query: 1016 DHEEDDIEHDD 1026
D ++DD E ++
Sbjct: 188 DDDDDDDEEEE 220
Score = 28.5 bits (62), Expect = 6.2
Identities = 22/63 (34%), Positives = 27/63 (41%)
Frame = +2
Query: 986 DADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
D DD+D GED G E D+ ++DD E + E EG EG G
Sbjct: 26 DEDDDDD------GEDDDGDEDDDDDAPGGGDDDDDEEDE------EEEGGVEG--GRGG 163
Query: 1046 GGD 1048
GGD
Sbjct: 164 GGD 172
Score = 28.1 bits (61), Expect = 8.1
Identities = 17/46 (36%), Positives = 22/46 (46%)
Frame = +2
Query: 1003 SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGD 1048
S + +GDE +D E+DD + DD D DA GGGD
Sbjct: 5 SRTRMSGDEDDDDDGEDDDGDEDDDD-------------DAPGGGD 103
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 43.5 bits (101), Expect = 2e-04
Identities = 29/107 (27%), Positives = 51/107 (47%), Gaps = 6/107 (5%)
Frame = -2
Query: 950 NAQSMAKSKHNIERRKYESRD---REEECGPETGGDNDADADDEDSENVSEAGEDVSGSE 1006
N +K H+++ ++ + + + E E+G + + D+ SE +S+ DV +
Sbjct: 548 NCVHQSKGHHDLQHQRVPNSNSS*KTESRYSESGDEGEEQGGDDGSEKLSD---DVDDTA 378
Query: 1007 SAGDECFQEDHEED---DIEHDDVDGKAESEGEAEGMCDAQGGGDSS 1050
GD E+ E D D+ DV A+ +G+ EG C GGGD +
Sbjct: 377 EEGDVTADEEAEGDGGVDVAAGDVG--ADGDGDEEGECVGDGGGDEA 243
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 40.8 bits (94), Expect = 0.001
Identities = 26/105 (24%), Positives = 51/105 (47%), Gaps = 5/105 (4%)
Frame = -2
Query: 927 EKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDAD 986
E ++G+ P G+ EED + + + + +K N +K GPE G+++ D
Sbjct: 587 EDDDGD-GPFGEGEEDLSSEDGAGYGNNSNNKSN--SKKAPEGGPGAGAGPEENGEDEED 417
Query: 987 ADDEDSENVSEAGEDVSGSESAGDE-----CFQEDHEEDDIEHDD 1026
D ED E+ + ED + G++ +ED+E+++ + +D
Sbjct: 416 EDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEED 282
>AV409241
Length = 282
Score = 37.4 bits (85), Expect = 0.013
Identities = 15/27 (55%), Positives = 18/27 (66%)
Frame = -1
Query: 21 ASSRGDSYGQSQVPGGGGGGGGAGGGG 47
A++ GD G + GGGGGGG GGGG
Sbjct: 183 AAAGGDGVGGGETVAGGGGGGGVGGGG 103
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 37.0 bits (84), Expect = 0.017
Identities = 23/71 (32%), Positives = 35/71 (48%)
Frame = +3
Query: 967 ESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDD 1026
++ + EEE E D D D DD+D +D G E D+ +D EE+ E +
Sbjct: 240 QTDNEEEEDDDEEEDDEDEDDDDDDD-------DDDDGEEEEDDD--DDDEEEEGSEEVE 392
Query: 1027 VDGKAESEGEA 1037
V+GK ++G A
Sbjct: 393 VEGKDGAKGAA 425
Score = 33.5 bits (75), Expect = 0.19
Identities = 18/64 (28%), Positives = 31/64 (48%)
Frame = +3
Query: 982 DNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMC 1041
DN+ + DD++ E +D + D+ + EE+D + DD + + E E EG
Sbjct: 246 DNEEEEDDDEEE------DDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKD 407
Query: 1042 DAQG 1045
A+G
Sbjct: 408 GAKG 419
>TC12258 weakly similar to UP|O50458 (O50458) PGRS_FAMILY protein (PE_PGRS
family protein), partial (6%)
Length = 471
Score = 36.6 bits (83), Expect = 0.023
Identities = 20/45 (44%), Positives = 23/45 (50%)
Frame = +3
Query: 20 FASSRGDSYGQSQVPGGGGGGGGAGGGGEATTSQKLTTNDALSYL 64
FASS G G S V GGGGGGG+GG + + DA L
Sbjct: 330 FASSAGGGNGGSGVGDGGGGGGGSGGESGDANLKLVGAGDAAQEL 464
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 35.0 bits (79), Expect = 0.066
Identities = 14/24 (58%), Positives = 17/24 (70%)
Frame = +1
Query: 24 RGDSYGQSQVPGGGGGGGGAGGGG 47
R + ++Q GGGGGGGG GGGG
Sbjct: 289 RNITVNEAQARGGGGGGGGRGGGG 360
Score = 30.0 bits (66), Expect = 2.1
Identities = 13/19 (68%), Positives = 14/19 (73%)
Frame = +1
Query: 29 GQSQVPGGGGGGGGAGGGG 47
G S+ GGGGGG G GGGG
Sbjct: 406 GGSRGYGGGGGGYGGGGGG 462
Score = 29.6 bits (65), Expect = 2.8
Identities = 13/23 (56%), Positives = 13/23 (56%)
Frame = +1
Query: 25 GDSYGQSQVPGGGGGGGGAGGGG 47
G G GGGGG GG GGGG
Sbjct: 397 GGGGGSRGYGGGGGGYGGGGGGG 465
Score = 29.3 bits (64), Expect = 3.6
Identities = 15/27 (55%), Positives = 16/27 (58%)
Frame = +1
Query: 21 ASSRGDSYGQSQVPGGGGGGGGAGGGG 47
A +RG G GGG GGGG GGGG
Sbjct: 310 AQARGGGGG-----GGGRGGGGYGGGG 375
>AV425102
Length = 426
Score = 35.0 bits (79), Expect = 0.066
Identities = 17/41 (41%), Positives = 23/41 (55%), Gaps = 1/41 (2%)
Frame = -1
Query: 19 PFASSRGDSYG-QSQVPGGGGGGGGAGGGGEATTSQKLTTN 58
PF D G ++++ GGGGGGGG GGG S ++ N
Sbjct: 138 PFGGVVADLRGYEAEISGGGGGGGGGGGGRRQ*KSDEVLQN 16
>TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor, partial
(92%)
Length = 1693
Score = 35.0 bits (79), Expect = 0.066
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 3/85 (3%)
Frame = +1
Query: 946 YRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGS 1005
Y A+ + + E+ +E +++ E E ++ AD+D ED E+ EA D +
Sbjct: 1132 YAKQVAEETWGKQKDAEKAAFEEAEKKRE--EEESKEDPADSDAEDEEDTDEASHDSDDA 1305
Query: 1006 ES---AGDECFQEDHEEDDIEHDDV 1027
ES AG++ D E+D+ HD++
Sbjct: 1306 ESKTEAGED--SADANEEDV-HDEL 1371
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein 1-like
protein 3, partial (81%)
Length = 1298
Score = 35.0 bits (79), Expect = 0.066
Identities = 21/61 (34%), Positives = 31/61 (50%)
Frame = +2
Query: 986 DADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
+A+ D E++ E ED GDE ED ++D+ E DD D E + E EG ++
Sbjct: 743 EAEQSDFEDIEEDDED-------GDEDEDEDDDDDEEEEDD-DDDDEDDEEGEGKSKSKS 898
Query: 1046 G 1046
G
Sbjct: 899 G 901
Score = 32.3 bits (72), Expect = 0.43
Identities = 28/135 (20%), Positives = 57/135 (41%), Gaps = 8/135 (5%)
Frame = +2
Query: 908 LVENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSK---HNI--- 961
+ + K +S P +V +++ ++ + E + D + S + K H +
Sbjct: 557 ITKTEKCESFFNFFNPPQVPEDDDDIDDDAVEELQNLMEHDYDIGSTIRDKIIPHAVSWF 736
Query: 962 --ERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEE 1019
E + + D EE+ + GD D D DD+D E +ED ++
Sbjct: 737 TGEAEQSDFEDIEED---DEDGDEDEDEDDDDDEE-------------------EEDDDD 850
Query: 1020 DDIEHDDVDGKAESE 1034
DD + ++ +GK++S+
Sbjct: 851 DDEDDEEGEGKSKSK 895
>BP034914
Length = 544
Score = 34.7 bits (78), Expect = 0.087
Identities = 21/72 (29%), Positives = 33/72 (45%)
Frame = +3
Query: 243 KQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKRDRRIRDQDERDPDLDNSRD 302
KQRY D PS S + D D + + +++R RR +R+ + R+
Sbjct: 39 KQRYVHDDTPSPSSSSSSSTAS-DSDSDSDRSSRRRREKERRRRSDGGSKRERESSRRRE 215
Query: 303 LTSQRFRDKKKT 314
+R RD+KKT
Sbjct: 216 KRKKRDRDRKKT 251
Score = 28.5 bits (62), Expect = 6.2
Identities = 26/120 (21%), Positives = 48/120 (39%), Gaps = 3/120 (2%)
Frame = +3
Query: 916 SHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEEC 975
S SS + + S E++ D ++ +S E+RK RDR++
Sbjct: 75 SSSSSSSTASDSDSDSDRSSRRRREKERRRRSDGGSKRERESSRRREKRKKRDRDRKKTK 254
Query: 976 GPETGGDNDADADDEDS---ENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAE 1032
D ++DED + +S E ++ + G++ Q DD + D+ G +E
Sbjct: 255 RHRHSDDESHSSEDEDKLRVQPLSVVRELMTEFPNVGNDLKQLLQMIDDGQAVDIKGISE 434
>TC12667 similar to PIR|D96525|D96525 protein T1N15.19 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(10%)
Length = 498
Score = 34.7 bits (78), Expect = 0.087
Identities = 19/38 (50%), Positives = 22/38 (57%), Gaps = 6/38 (15%)
Frame = -1
Query: 21 ASSRGDSYGQSQ--VPGGGGG----GGGAGGGGEATTS 52
A GD +G V GGGGG GGG GGGGEA+ +
Sbjct: 348 AVDEGDPFGPGMELVGGGGGGCDFFGGGGGGGGEASAA 235
>TC11625 weakly similar to UP|O81015 (O81015) At2g26920 protein, partial
(8%)
Length = 410
Score = 34.7 bits (78), Expect = 0.087
Identities = 14/18 (77%), Positives = 15/18 (82%)
Frame = +2
Query: 30 QSQVPGGGGGGGGAGGGG 47
+S V GGGGGGGG GGGG
Sbjct: 245 RSVVDGGGGGGGGGGGGG 298
Score = 32.7 bits (73), Expect = 0.33
Identities = 15/26 (57%), Positives = 16/26 (60%)
Frame = +2
Query: 25 GDSYGQSQVPGGGGGGGGAGGGGEAT 50
G S + V GGGGGGG GGGG T
Sbjct: 227 GVSERERSVVDGGGGGGGGGGGGGIT 304
Score = 28.5 bits (62), Expect = 6.2
Identities = 11/15 (73%), Positives = 11/15 (73%)
Frame = +2
Query: 37 GGGGGGAGGGGEATT 51
GGGGGG GGGG T
Sbjct: 260 GGGGGGGGGGGGGIT 304
>BP043764
Length = 479
Score = 34.7 bits (78), Expect = 0.087
Identities = 13/15 (86%), Positives = 14/15 (92%)
Frame = -2
Query: 35 GGGGGGGGAGGGGEA 49
GGGGGGGG GGGGE+
Sbjct: 349 GGGGGGGGGGGGGES 305
Score = 33.5 bits (75), Expect = 0.19
Identities = 14/21 (66%), Positives = 14/21 (66%)
Frame = -2
Query: 35 GGGGGGGGAGGGGEATTSQKL 55
GGGGGGGG GGG SQ L
Sbjct: 346 GGGGGGGGGGGGESLLVSQSL 284
Score = 32.0 bits (71), Expect = 0.56
Identities = 16/40 (40%), Positives = 20/40 (50%)
Frame = -2
Query: 8 IYSASASQFKRPFASSRGDSYGQSQVPGGGGGGGGAGGGG 47
I+ ++F S S +S GGGGGGG GGGG
Sbjct: 433 IFPGCRNRFTSSAPLSSAISVSRSGGGNGGGGGGGGGGGG 314
Score = 31.2 bits (69), Expect = 0.96
Identities = 17/42 (40%), Positives = 20/42 (47%)
Frame = -2
Query: 20 FASSRGDSYGQSQVPGGGGGGGGAGGGGEATTSQKLTTNDAL 61
F SS S S GGG GGG GGGG + L + +L
Sbjct: 409 FTSSAPLSSAISVSRSGGGNGGGGGGGGGGGGGESLLVSQSL 284
>TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (4%)
Length = 493
Score = 34.3 bits (77), Expect = 0.11
Identities = 28/79 (35%), Positives = 38/79 (47%), Gaps = 1/79 (1%)
Frame = -2
Query: 969 RDREEECGPETGGDND-ADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDV 1027
R +E G DND A A++E+ E E GE+ + DE +ED EED H V
Sbjct: 438 RGEDETVIDGAGEDNDRAVAEEEEEEPGEEDGEEDDDGDGVPDEGDEEDEEED---HGVV 268
Query: 1028 DGKAESEGEAEGMCDAQGG 1046
+E AE + A+GG
Sbjct: 267 -----AEEVAEVVLHAEGG 226
Score = 28.5 bits (62), Expect = 6.2
Identities = 22/71 (30%), Positives = 31/71 (42%), Gaps = 2/71 (2%)
Frame = -2
Query: 970 DREEECGPETGGDNDADADDEDSENVSEAGEDVSGS--ESAGDECFQEDHEEDDIEHDDV 1027
D E G + + ED + AGED + E +E +ED EEDD + D V
Sbjct: 492 DIEPNSGENERNNGNNPERGEDETVIDGAGEDNDRAVAEEEEEEPGEEDGEEDD-DGDGV 316
Query: 1028 DGKAESEGEAE 1038
+ + E E E
Sbjct: 315 PDEGDEEDEEE 283
>TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1
(AT3g17100/K14A17_22_), partial (43%)
Length = 1108
Score = 34.3 bits (77), Expect = 0.11
Identities = 13/18 (72%), Positives = 15/18 (83%)
Frame = -3
Query: 30 QSQVPGGGGGGGGAGGGG 47
+S+ GGGGGGGG GGGG
Sbjct: 62 ESEEKGGGGGGGGGGGGG 9
>TC19505 weakly similar to UP|Q9FG35 (Q9FG35) Emb|CAB82953.1, partial (12%)
Length = 723
Score = 34.3 bits (77), Expect = 0.11
Identities = 13/14 (92%), Positives = 13/14 (92%)
Frame = -2
Query: 35 GGGGGGGGAGGGGE 48
GGGGGGGG GGGGE
Sbjct: 473 GGGGGGGGGGGGGE 432
Score = 32.3 bits (72), Expect = 0.43
Identities = 12/13 (92%), Positives = 12/13 (92%)
Frame = -2
Query: 35 GGGGGGGGAGGGG 47
GGGGGGGG GGGG
Sbjct: 476 GGGGGGGGGGGGG 438
Score = 31.2 bits (69), Expect = 0.96
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = -2
Query: 25 GDSYGQSQVPGGGGGGGGAGGGG 47
G++ Q + G GGGGGG GGGG
Sbjct: 512 GENGRQLRFRGTGGGGGGGGGGG 444
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,115,549
Number of Sequences: 28460
Number of extensions: 203526
Number of successful extensions: 3824
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 1741
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2661
length of query: 1129
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1029
effective length of database: 2,051,600
effective search space: 2111096400
effective search space used: 2111096400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0081b.6


