

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.3
(322 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
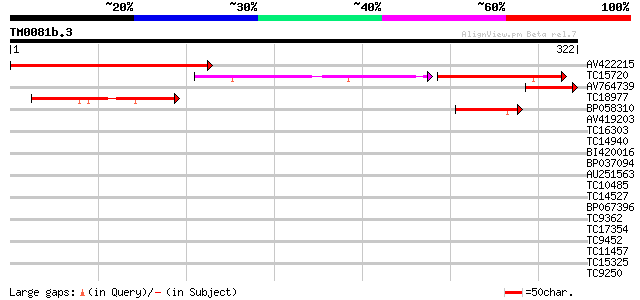
Score E
Sequences producing significant alignments: (bits) Value
AV422215 236 4e-63
TC15720 similar to UP|O35328 (O35328) Proline-rich protein 9-1 (... 98 2e-42
AV764739 69 1e-12
TC18977 similar to AAQ62440 (AAQ62440) At1g74870, partial (5%) 69 1e-12
BP058310 45 1e-05
AV419203 35 0.022
TC16303 weakly similar to UP|AGA1_YEAST (P32323) A-agglutinin at... 34 0.029
TC14940 34 0.038
BI420016 33 0.065
BP037094 33 0.065
AU251563 32 0.19
TC10485 32 0.19
TC14527 similar to PIR|F86340|F86340 protein F2D10.34 [imported]... 32 0.19
BP067396 31 0.25
TC9362 similar to PIR|F86340|F86340 protein F2D10.34 [imported] ... 31 0.32
TC17354 weakly similar to UP|Q9LS15 (Q9LS15) Gb|AAF26010.1, part... 31 0.32
TC9452 similar to UP|O04259 (O04259) Drought-induced-19-like 1 (... 30 0.55
TC11457 30 0.55
TC15325 similar to UP|O82268 (O82268) At2g31160 protein (At2g311... 30 0.72
TC9250 UP|Q9FLP6 (Q9FLP6) Ubiquitin-like protein SMT3-like (AT5g... 30 0.72
>AV422215
Length = 409
Score = 236 bits (602), Expect = 4e-63
Identities = 115/115 (100%), Positives = 115/115 (100%)
Frame = +1
Query: 1 MASGFISDPPLSKKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSNGRMDNCPSSKHT 60
MASGFISDPPLSKKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSNGRMDNCPSSKHT
Sbjct: 64 MASGFISDPPLSKKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSNGRMDNCPSSKHT 243
Query: 61 ASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSS 115
ASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSS
Sbjct: 244 ASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSS 408
>TC15720 similar to UP|O35328 (O35328) Proline-rich protein 9-1 (Fragment),
partial (11%)
Length = 1115
Score = 97.8 bits (242), Expect(2) = 2e-42
Identities = 43/75 (57%), Positives = 53/75 (70%), Gaps = 2/75 (2%)
Frame = +1
Query: 244 SQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKLYDH--VDGNV 301
S PS CPIC+EDLD+TD+SFLPC CGF LCLFCHK+ILE D RCP CRK Y+ V+
Sbjct: 502 SAPSSCPICFEDLDLTDTSFLPCLCGFRLCLFCHKRILEDDGRCPGCRKPYEREPVETEA 681
Query: 302 GLNIGAKAFCITQSC 316
+ G+ + +SC
Sbjct: 682 SVVGGSLTLRLARSC 726
Score = 91.7 bits (226), Expect(2) = 2e-42
Identities = 61/154 (39%), Positives = 77/154 (49%), Gaps = 19/154 (12%)
Frame = +3
Query: 106 SSGKDFSGSSSNSISTSCSG-------------NDSEEEDDDGCLDDWEAVADALYANDK 152
+ G GSSS+SIS+S +G + EEE DD LDDWEAVADAL A+DK
Sbjct: 63 TGGSSGGGSSSSSISSSSAGCFSGYITEVDEEVEEEEEEGDDEGLDDWEAVADALAADDK 242
Query: 153 SHSVVSESPAEHEAECRYTVLEDDKNPRVDFSKADFKSE------VPESKPNRRAWKPDD 206
+ S+ P H+ TV PR + +D S+ VP N RAW+ DD
Sbjct: 243 NQCPCSDLPPSHDEHVGQTV-----PPRAAANGSDSNSKPASGRLVPWGSGNVRAWRADD 407
Query: 207 TLRPRCLPNLSKQRNSPLNSNWRGSRKTVPWAWQ 240
RP+ LPNLSKQ + P + G VPW Q
Sbjct: 408 AFRPQSLPNLSKQHSMPNPDRYCGG---VPWCSQ 500
>AV764739
Length = 338
Score = 68.6 bits (166), Expect = 1e-12
Identities = 29/29 (100%), Positives = 29/29 (100%)
Frame = -3
Query: 294 YDHVDGNVGLNIGAKAFCITQSCRTGTGW 322
YDHVDGNVGLNIGAKAFCITQSCRTGTGW
Sbjct: 336 YDHVDGNVGLNIGAKAFCITQSCRTGTGW 250
>TC18977 similar to AAQ62440 (AAQ62440) At1g74870, partial (5%)
Length = 449
Score = 68.6 bits (166), Expect = 1e-12
Identities = 43/91 (47%), Positives = 56/91 (61%), Gaps = 7/91 (7%)
Frame = +2
Query: 13 KKKRTNGSAKLKQIKLDVRREQWLSR---VKKGC--NVDSNGRMDNCPSSKHTASEENRS 67
KKKRTN SAKLKQ K+D RREQWL++ KGC VD +G P+ A ++ +
Sbjct: 167 KKKRTNRSAKLKQYKIDARREQWLNQGAVKSKGCKDGVDDDGHALPSPA----AGKQGKG 334
Query: 68 SCK--EMTRKGEDIEGTCIQDSDSRSTINSP 96
S + EM R+G++ +G QDSDS S NSP
Sbjct: 335 SLQQLEMRREGDEDDGLIHQDSDSESPSNSP 427
>BP058310
Length = 480
Score = 45.4 bits (106), Expect = 1e-05
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 6/44 (13%)
Frame = +1
Query: 254 EDLDVTDSSFLPCSCGFHLCLFCHKKIL------EADARCPSCR 291
E++D+TD PC CG+ +C++C I+ + + RCP+CR
Sbjct: 139 EEMDLTDQQLKPCKCGYEICVWCWHHIMDMAEKDDTEGRCPACR 270
>AV419203
Length = 393
Score = 34.7 bits (78), Expect = 0.022
Identities = 16/46 (34%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Frame = +1
Query: 247 SQCPICYEDL-DVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCR 291
S+CP+C E+ + + LPC+ FH+ C + L + +CP CR
Sbjct: 250 SECPVCLEEFHEGNEVRGLPCAHNFHV--ECIDEWLRLNVKCPRCR 381
>TC16303 weakly similar to UP|AGA1_YEAST (P32323) A-agglutinin attachment
subunit precursor, partial (3%)
Length = 1235
Score = 34.3 bits (77), Expect = 0.029
Identities = 36/135 (26%), Positives = 48/135 (34%), Gaps = 7/135 (5%)
Frame = +1
Query: 50 RMDNCPSSKHTASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGK 109
R D K ++ S KE KG+ G+ S S + +GK
Sbjct: 394 RSDEGEGRKRKKEKDGGSGKKEKRLKGDTRFGSSSSGGKGGSKFGSAKKG---FGGKAGK 564
Query: 110 DFSGSSSNSISTSCSGNDSEEE-------DDDGCLDDWEAVADALYANDKSHSVVSESPA 162
D G T +G DSE++ DDD +DD V Y N S E+P
Sbjct: 565 DHDGEVKEMWDT-IAGGDSEDDQEGVRNLDDDNFIDD-TGVEPGFYGNYNEPSSPGEAPQ 738
Query: 163 EHEAECRYTVLEDDK 177
E E EDD+
Sbjct: 739 AEEGE------EDDE 765
>TC14940
Length = 781
Score = 33.9 bits (76), Expect = 0.038
Identities = 17/44 (38%), Positives = 21/44 (47%)
Frame = +2
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
CPIC L S+ CG C C K + A +CP+CRK
Sbjct: 149 CPICMGPLVEEMST----RCGHIFCKACIKAAISAQNKCPTCRK 268
>BI420016
Length = 559
Score = 33.1 bits (74), Expect = 0.065
Identities = 22/68 (32%), Positives = 34/68 (49%)
Frame = +1
Query: 112 SGSSSNSISTSCSGNDSEEEDDDGCLDDWEAVADALYANDKSHSVVSESPAEHEAECRYT 171
S SSSNS S S +G + ++ DD WE+V+ A + HS+ + P H + +
Sbjct: 118 SPSSSNSSSMSAAGTPNPQKTDDPMHSWWESVSK---ARSRIHSLATILPNSHSS--ALS 282
Query: 172 VLEDDKNP 179
L D + P
Sbjct: 283 SLADSERP 306
>BP037094
Length = 502
Score = 33.1 bits (74), Expect = 0.065
Identities = 19/51 (37%), Positives = 24/51 (46%), Gaps = 1/51 (1%)
Frame = -3
Query: 244 SQPSQCPICYEDL-DVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKL 293
S S C IC + D FLP C H + C K L + + CP+CR L
Sbjct: 452 SSGSSCVICLAEFCDGEQIRFLP-KCNHHFHVVCIDKWLLSHSSCPTCRNL 303
>AU251563
Length = 350
Score = 31.6 bits (70), Expect = 0.19
Identities = 25/92 (27%), Positives = 36/92 (38%)
Frame = +2
Query: 50 RMDNCPSSKHTASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGK 109
R PS H+AS SS + ED + S++S D G
Sbjct: 32 RSSQSPSPSHSASASATSSIHKRKLVSED-------------HVPPFPPSSFSADTRDGA 172
Query: 110 DFSGSSSNSISTSCSGNDSEEEDDDGCLDDWE 141
S SIS + +DS+E+ +D +DD E
Sbjct: 173 LTSNDDLESISARGADSDSDEDSEDAVVDDEE 268
>TC10485
Length = 636
Score = 31.6 bits (70), Expect = 0.19
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = -1
Query: 105 ESSGKDFSGSSSNSISTSCSGNDSEEEDDDGCLDDW 140
+S GK S SSS+S +D +++DD+G W
Sbjct: 315 KSGGKQLSSSSSSSREVCVFSSDDDDDDDEGSCASW 208
>TC14527 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(42%)
Length = 1095
Score = 31.6 bits (70), Expect = 0.19
Identities = 14/47 (29%), Positives = 23/47 (48%)
Frame = +2
Query: 247 SQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKL 293
++C IC + D D + CG + C L + + CPSCR++
Sbjct: 503 TECAICLAEFDAGDEIRVLPQCGHGFHVGCIDTWLGSHSSCPSCRQV 643
>BP067396
Length = 506
Score = 31.2 bits (69), Expect = 0.25
Identities = 24/81 (29%), Positives = 33/81 (40%), Gaps = 8/81 (9%)
Frame = -1
Query: 244 SQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKLYDHVDGNV-- 301
S S C IC D D+ + CG L C L+ + CP CRK V ++
Sbjct: 401 SVSSCCSICLGDYKEADTLRMLPDCGHVYHLACVDPWLKFHSTCPICRK--SSVQSSIKV 228
Query: 302 ----GLNIG--AKAFCITQSC 316
NIG + FC+ +C
Sbjct: 227 *LVSAYNIGEPCETFCV*HAC 165
>TC9362 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(58%)
Length = 591
Score = 30.8 bits (68), Expect = 0.32
Identities = 14/47 (29%), Positives = 22/47 (46%)
Frame = +1
Query: 247 SQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKL 293
S C IC + D + CG + C L++ + CPSCR++
Sbjct: 322 SDCAICLTEFAAGDEIRVLPQCGHGFHVSCIDAWLKSHSSCPSCRQI 462
>TC17354 weakly similar to UP|Q9LS15 (Q9LS15) Gb|AAF26010.1, partial (24%)
Length = 638
Score = 30.8 bits (68), Expect = 0.32
Identities = 21/49 (42%), Positives = 27/49 (54%), Gaps = 4/49 (8%)
Frame = +3
Query: 90 RSTINSPDR----STYSHDESSGKDFSGSSSNSISTSCSGNDSEEEDDD 134
R+ +N+ R ST S SG SGS S S S+S S +DSE D+D
Sbjct: 180 RAQVNAEGRDEQTSTSSSSSGSGSSESGSGSGSRSSS-SSSDSEGSDED 323
>TC9452 similar to UP|O04259 (O04259) Drought-induced-19-like 1 (T2H3.13
protein), partial (12%)
Length = 570
Score = 30.0 bits (66), Expect = 0.55
Identities = 16/49 (32%), Positives = 25/49 (50%)
Frame = -2
Query: 88 DSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTSCSGNDSEEEDDDGC 136
D +S + R +S SGKD S S SN++S+ C + ++ GC
Sbjct: 158 DIKSPCTNLLRRAFSMRSWSGKDGSTSLSNTVSSECVSSMLNSLEESGC 12
>TC11457
Length = 629
Score = 30.0 bits (66), Expect = 0.55
Identities = 18/51 (35%), Positives = 25/51 (48%), Gaps = 2/51 (3%)
Frame = +1
Query: 247 SQCPICYEDLDVTD-SSFLP-CSCGFHLCLFCHKKILEADARCPSCRKLYD 295
S+C +C +++D + LP C+ GFH L C L CP CR D
Sbjct: 376 SECAVCLDEIDSDQPARVLPGCNHGFH--LECADTWLSKHPLCPVCRAKLD 522
>TC15325 similar to UP|O82268 (O82268) At2g31160 protein
(At2g31160/T16B12.3), partial (32%)
Length = 645
Score = 29.6 bits (65), Expect = 0.72
Identities = 20/72 (27%), Positives = 34/72 (46%)
Frame = +2
Query: 62 SEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSIST 121
S + + S K + ++ GTC DS T N+ + S SG S S+SNS +
Sbjct: 275 SAKVQGSVKVLMDSIQEFIGTCNSDSTCSLTNNTSMTTLTSLASGSGASSSPSASNSTAA 454
Query: 122 SCSGNDSEEEDD 133
+ S ++++ D
Sbjct: 455 TSSRYENQKRRD 490
>TC9250 UP|Q9FLP6 (Q9FLP6) Ubiquitin-like protein SMT3-like
(AT5g55160/MCO15_11) (Small ubiquitin-like modifier 2),
partial (22%)
Length = 1363
Score = 29.6 bits (65), Expect = 0.72
Identities = 15/44 (34%), Positives = 18/44 (40%)
Frame = +1
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
CP C L T S SCG C C + + +D C C K
Sbjct: 952 CPSCKVTLTNTMSLVAISSCGHVFCKKCADRFMASDKVCLVCNK 1083
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.129 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,693,763
Number of Sequences: 28460
Number of extensions: 105725
Number of successful extensions: 653
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 633
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 645
length of query: 322
length of database: 4,897,600
effective HSP length: 90
effective length of query: 232
effective length of database: 2,336,200
effective search space: 541998400
effective search space used: 541998400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0081b.3


