

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076a.9
(388 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
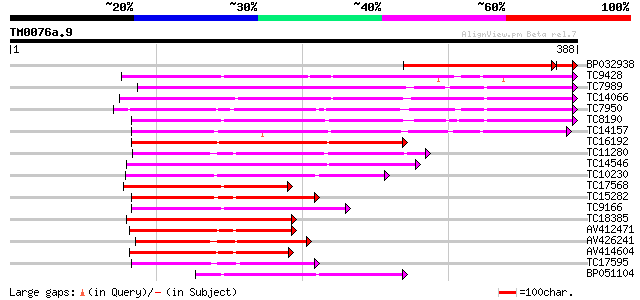
Score E
Sequences producing significant alignments: (bits) Value
BP032938 218 3e-61
TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), pa... 198 1e-51
TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , par... 180 4e-46
TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 ,... 178 2e-45
TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), part... 166 5e-42
TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 p... 162 7e-41
TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , p... 160 4e-40
TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%) 159 7e-40
TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), p... 152 7e-38
TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxida... 124 3e-29
TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precurso... 119 9e-28
TC17568 similar to UP|Q8RVP6 (Q8RVP6) Gaiacol peroxidase , part... 112 1e-25
TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fra... 108 2e-24
TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%) 101 2e-22
TC18385 similar to UP|Q9XFL6 (Q9XFL6) Peroxidase 5, partial (38%) 100 3e-22
AV412471 99 1e-21
AV426241 98 2e-21
AV414604 97 3e-21
TC17595 similar to UP|Q9XFI7 (Q9XFI7) Peroxidase (Fragment), par... 97 5e-21
BP051104 94 5e-20
>BP032938
Length = 475
Score = 218 bits (556), Expect(2) = 3e-61
Identities = 105/105 (100%), Positives = 105/105 (100%)
Frame = -2
Query: 270 FKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLVDKGKSL 329
FKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLVDKGKSL
Sbjct: 474 FKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLVDKGKSL 295
Query: 330 LLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLT 374
LLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLT
Sbjct: 294 LLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLT 160
Score = 33.9 bits (76), Expect(2) = 3e-61
Identities = 14/14 (100%), Positives = 14/14 (100%)
Frame = -3
Query: 375 GNDGEVRKICRATN 388
GNDGEVRKICRATN
Sbjct: 158 GNDGEVRKICRATN 117
>TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1354
Score = 198 bits (503), Expect = 1e-51
Identities = 114/317 (35%), Positives = 176/317 (54%), Gaps = 5/317 (1%)
Frame = +2
Query: 77 PNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASIL 136
P + + L FY D+CPN I+ + + +++P+ + +++RL FHDCFV GCDASIL
Sbjct: 95 PYSSDAQLDNSFYKDTCPNVHSIVREVVRNVSKSDPRMLGSLIRLHFHDCFVQGCDASIL 274
Query: 137 LDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA- 195
L+ T + E+ +F N ++G D+V+ IK+ +E CP VSCAD + + + LA
Sbjct: 275 LNDTSTIVS-EQGAFPNNNSIRGLDVVNQIKTAVENACPNTVSCADILALAAEISSVLAN 451
Query: 196 GMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSI 255
G+ + PLG RRDSL + ++ + N LP PN++ ++ F + + ++V L GAH+I
Sbjct: 452 GVDWKVPLG-RRDSLTANRSLANQN-LPGPNFNLSQLNASFAVQNLNITDLVALSGAHTI 625
Query: 256 GAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQIC--SNPGTPRYRNEPVNFDNTPTV 313
G C F++R+YNF NT PDP L +L LR IC PGT +P TP
Sbjct: 626 GRGQCQFFVNRLYNFSNTGNPDPTLNTTYLQTLRSICPHGGPGTTLAHLDP----TTPDT 793
Query: 314 MDNLFYRDLVDKGKSLLLTDAHL--VTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLN 371
+D+ ++ +L GK L +D L + T V + + LF + F M K++ +
Sbjct: 794 LDSQYFSNL-QGGKGLFQSDPVLFSTSGAATVAIVNSFSSNPTLFFENFKASMIKMSRIG 970
Query: 372 VLTGNDGEVRKICRATN 388
VLTG+ GE+RK C N
Sbjct: 971 VLTGSQGEIRKPCNFVN 1021
>TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , partial (85%)
Length = 1352
Score = 180 bits (456), Expect = 4e-46
Identities = 105/301 (34%), Positives = 157/301 (51%)
Frame = +3
Query: 88 FYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNVE 147
FY SCP E II ++ + A +LRL FHDCFV GCDAS+LLD + +G + +
Sbjct: 150 FYDSSCPKLESIIRKELKKVFDKDIAQAAGLLRLHFHDCFVQGCDASVLLDGSASGPSEK 329
Query: 148 KSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLGGRR 207
+ + + +++D++S++E++C +VSCAD + +A+ L+G P + GRR
Sbjct: 330 DAPPNLTLRAQAFKIIEDLRSQIEKKCGRVVSCADITAIAARDAVFLSGGPDYELPLGRR 509
Query: 208 DSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIFMDRV 267
D L V +NLP P + ++ K P ++V L G H+IG +HC F DR+
Sbjct: 510 DGLNFATRNVTLDNLPAPQSNTTTILNSLATKNLDPTDVVSLSGGHTIGISHCSSFTDRL 689
Query: 268 YNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLVDKGK 327
Y K DP + F L+ C T N V +P DN +Y DL+++ +
Sbjct: 690 YPSK-----DPVMDQTFEKNLKLTCPASNTD---NTTVLDLRSPNTFDNKYYVDLMNR-Q 842
Query: 328 SLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRKICRAT 387
L +D L TD RT V A +Q+LF ++F M K+ LNVLTG+ GE+R C
Sbjct: 843 GLFFSDQDLYTDKRTKDIVTSFAVNQSLFFEKFVVAMLKMGQLNVLTGSQGEIRANCSVR 1022
Query: 388 N 388
N
Sbjct: 1023N 1025
>TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(92%)
Length = 1436
Score = 178 bits (451), Expect = 2e-45
Identities = 114/314 (36%), Positives = 162/314 (51%), Gaps = 1/314 (0%)
Frame = +2
Query: 76 QPNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASI 135
Q + + L FY ++CP AE II + + + + + LR FHDC V CDAS+
Sbjct: 128 QEDVQDTGLAMNFYKETCPQAEDIITEQVKLLYKRHKNTAFSWLRNIFHDCAVQSCDASL 307
Query: 136 LLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA 195
LLDST + +++ G L+ ++ IK LE ECPG+VSC+D +V + + +A
Sbjct: 308 LLDSTRKTLSEKETDRSFG--LRNFRYIETIKEALERECPGVVSCSDILVLSARDGIAAL 481
Query: 196 GMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSI 255
G P GRRD S A VV+ LP N S +++ F G +V LLGAHS+
Sbjct: 482 GGPHIPLRTGRRDGRRSRADVVEQF-LPDHNESISAVLDKFGAMGIDTPGVVALLGAHSV 658
Query: 256 GAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNP-GTPRYRNEPVNFDNTPTVM 314
G HC + R+Y + DPAL P + + + C + P+ N TP ++
Sbjct: 659 GRTHCVKLVHRLYP-----EVDPALNPDHIPHMLKKCPDAIPDPKAVQYVRNDRGTPMIL 823
Query: 315 DNLFYRDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLT 374
DN +YR+++D K LLL D L D RT P V +MA Q F K F+ +T L+ N LT
Sbjct: 824 DNNYYRNILDN-KGLLLVDHQLANDKRTKPYVKKMAKSQDYFFKEFSRAITLLSENNPLT 1000
Query: 375 GNDGEVRKICRATN 388
G GE+RK C N
Sbjct: 1001GTKGEIRKQCNVAN 1042
>TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), partial (86%)
Length = 1286
Score = 166 bits (421), Expect = 5e-42
Identities = 103/317 (32%), Positives = 165/317 (51%)
Frame = +1
Query: 72 EFQDQPNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGC 131
E Q +P + L FYS +CP E ++ + ++++ + +LR+ FHDCFV GC
Sbjct: 73 EAQTKPPVVE-GLSFSFYSKTCPKLETVVRNHLKKVLKKDNGQAPGLLRIFFHDCFVQGC 249
Query: 132 DASILLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEA 191
D S+LLD +P G+ + ++ GI + ++DI++ + ++C IVSCAD + S +A
Sbjct: 250 DGSVLLDGSP-GERDQPANI--GIRPEALQTIEDIRALVHKQCGKIVSCADITILASRDA 420
Query: 192 MALAGMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLG 251
+ L G P GRRD + S +T V LP P + ++ F + F ++V L G
Sbjct: 421 VFLTGGPDYAVPLGRRDGV-SFST-VGTQKLPSPINNTTATLKAFADRNFDATDVVALSG 594
Query: 252 AHSIGAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTP 311
AH+ G AHC F +R+ + DP + L C + N + TP
Sbjct: 595 AHTFGRAHCGTFFNRL------SPLDPNMDKTLAKNLTATCPAQNSTNTANLDI---RTP 747
Query: 312 TVMDNLFYRDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLN 371
V DN +Y DL+++ + + +D L++D RT V A +Q LF ++F + + KL+ L+
Sbjct: 748 NVFDNKYYLDLMNR-QGVFTSDQDLLSDKRTKGLVNAFAVNQTLFFEKFVDAVIKLSQLD 924
Query: 372 VLTGNDGEVRKICRATN 388
VLTGN GE+R C N
Sbjct: 925 VLTGNQGEIRGRCNVVN 975
>TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 precursor
(PNPC1) , partial (96%)
Length = 1202
Score = 162 bits (411), Expect = 7e-41
Identities = 110/306 (35%), Positives = 158/306 (50%), Gaps = 1/306 (0%)
Frame = +1
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L FY+ +CP I V + A++LRL FHDCFV GCDASILLD T +
Sbjct: 142 LSSTFYAKTCPLVLATIKTQVNLAVAKEARMGASLLRLHFHDCFVQGCDASILLDDTSSF 321
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEA-MALAGMPRQKP 202
EK++ N ++G D++D IKSK+E CPG+VSCAD + + ++ +AL G P
Sbjct: 322 TG-EKTAGPNANSVRGYDVIDTIKSKVESLCPGVVSCADIVAVAARDSVVALGGFSWAVP 498
Query: 203 LGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDI 262
LG R + SL++ ++ LP P+ + D + F KGF+ EMV L G+H+IG A C
Sbjct: 499 LGRRDSTTASLSSA--NSELPGPSSNLDGLNTAFSNKGFTTREMVALSGSHTIGQARCLF 672
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDL 322
F R+Y + + F L+ C G P++ +PT DN +YR+L
Sbjct: 673 FRTRIYY-------ETHIDSTFAKNLQGNCPFNGGDS-NLSPLD-TTSPTTFDNGYYRNL 825
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRK 382
+ K L +D L T V + A F FA M K+ +L+ LTG+ G++R
Sbjct: 826 QSQ-KGLFHSDQVLFNGGSTDSQVNSYVTNPASFKTDFANAMVKMGNLSPLTGSSGQIRT 1002
Query: 383 ICRATN 388
CR TN
Sbjct: 1003HCRKTN 1020
>TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , partial
(96%)
Length = 1390
Score = 160 bits (404), Expect = 4e-40
Identities = 100/306 (32%), Positives = 158/306 (50%), Gaps = 5/306 (1%)
Frame = +1
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L Y+ +CPN + I+ + + + LRL FHDCFV GCDAS+++ S+ G
Sbjct: 127 LSPNHYASTCPNLQSIVKGVVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVMVASS--G 300
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLE----EECPGIVSCADTMVFLSFEAMALAGMPR 199
+N + + + L G IK+K +C VSCAD + + + + LAG P
Sbjct: 301 NNKAEKDHPDNLSLAGDGFDTVIKAKAAVDAVPQCRNKVSCADILALATRDVVVLAGGPS 480
Query: 200 QKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAH 259
GR D L S A+ V+ LP PN++ +++ LF +G + +M+ L GAH++G +H
Sbjct: 481 YTVELGRFDGLVSRASDVN-GRLPEPNFNLNQLNSLFASQGLTQTDMIALSGAHTLGFSH 657
Query: 260 CDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNLF 318
C+ F +R+Y + DP L + +L+Q+C P+ +N D TP DN++
Sbjct: 658 CNRFSNRIY----STPVDPTLNRNYATQLQQMCPKNVNPQI---AINMDPTTPRTFDNIY 816
Query: 319 YRDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDG 378
Y++L +GK L +D L TD R+ TV A + F+ FA M KL + V T +G
Sbjct: 817 YKNL-QQGKGLFTSDQILFTDQRSKATVNSFASNSNTFNANFAAAMIKLGRVGVKTARNG 993
Query: 379 EVRKIC 384
++R C
Sbjct: 994 KIRTDC 1011
>TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%)
Length = 645
Score = 159 bits (402), Expect = 7e-40
Identities = 84/189 (44%), Positives = 118/189 (61%)
Frame = +2
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L G+YS SC AE I+ D V NP A ++R+ FHDCF+ GCDAS+LLDSTP+
Sbjct: 83 LEVGYYSYSCGMAEFIVKDEVRRSVTKNPGIAAGLVRMHFHDCFIRGCDASVLLDSTPS- 259
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
+ EK S N L+G +++D K+KLE C G+VSCAD + F + +++ LAG
Sbjct: 260 NTAEKDSPANKPSLRGFEVIDSAKAKLEAVCKGVVSCADIIAFAARDSVELAGGVGYDVP 439
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GRRD SLA+ +LP P ++ +++ +LF +KG + EEMV L GAH+IG +HC F
Sbjct: 440 AGRRDGRISLASDT-RTDLPPPTFNVNQLTQLFAKKGLTQEEMVTLSGAHTIGRSHCAAF 616
Query: 264 MDRVYNFKN 272
R+YNF +
Sbjct: 617 SSRLYNFSS 643
>TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), partial
(68%)
Length = 614
Score = 152 bits (385), Expect = 7e-38
Identities = 82/204 (40%), Positives = 122/204 (59%)
Frame = +3
Query: 85 REGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGD 144
R GFY +CP AE I+ V ++P A +LR+ FHDCFV GCDAS+L+ G
Sbjct: 27 RVGFYLGTCPRAESIVRSTVESHVNSDPTLAAGLLRMHFHDCFVQGCDASVLI----AGA 194
Query: 145 NVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLG 204
E+++ N + L+G +++DD K+K+E CPG+VSCAD + + +++ L+G +
Sbjct: 195 GTERTAIPN-LSLRGFEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPT 371
Query: 205 GRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIFM 264
GRRD S A+ D NNLP P S D + F KG + +++V L+G H+IG C F
Sbjct: 372 GRRDGRVSQAS--DVNNLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGGHTIGTTACQFFS 545
Query: 265 DRVYNFKNTNKPDPALRPPFLNEL 288
+R+YNF +N PDP++ FL +L
Sbjct: 546 NRLYNF-TSNGPDPSIDASFLLQL 614
>TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxidase
precursor , partial (66%)
Length = 738
Score = 124 bits (310), Expect = 3e-29
Identities = 69/201 (34%), Positives = 104/201 (51%)
Frame = +1
Query: 81 NPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDST 140
N L FY SCP + I+ +A + ++ + A+ILRL FHDCFV GCD SILLD
Sbjct: 91 NAQLTPTFYDRSCPKLQTIVRNAMVQTIKKEARMGASILRLFFHDCFVNGCDGSILLDDI 270
Query: 141 PNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQ 200
EK++ N +G +++D IK+ +E C VSCAD + + + + L G P
Sbjct: 271 GTTFVGEKNAAPNKNSARGFEVIDTIKTNVEASCNNTVSCADILALATRDGINLLGGPTW 450
Query: 201 KPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHC 260
+ GRRD+ + + + +P P+ ++ +F KG S ++ +L G H+IG A C
Sbjct: 451 QVPLGRRDARTASQSKA-NTEIPSPSSDLSTLISMFSAKGLSARDLTVLSGGHTIGQAEC 627
Query: 261 DIFMDRVYNFKNTNKPDPALR 281
F RV N N + A R
Sbjct: 628 QFFRSRVNNETNIDAAFAASR 690
>TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precursor (Atperox
P10) (ATP5a) , partial (42%)
Length = 753
Score = 119 bits (298), Expect = 9e-28
Identities = 66/181 (36%), Positives = 100/181 (54%)
Frame = +3
Query: 80 INPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDS 139
++ L FY +CPN +I+ + + + A++LRL FHDCFV GCD S+LLD
Sbjct: 213 VSSQLYYNFYDTTCPNLTRIVRYNLLSAMSNDTRIAASLLRLHFHDCFVNGCDGSVLLDD 392
Query: 140 TPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPR 199
T + EK++ N ++G +++D IKS LE+ CP VSCAD + + EA+ L+ P
Sbjct: 393 T-STQKGEKNALPNKNSIRGFEVIDTIKSALEKACPSTVSCADILTLAAREAVYLSRGPF 569
Query: 200 QKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAH 259
GRRD + A+ + NNLP P + + F KG +++ +L GAH+ G A
Sbjct: 570 WSVPLGRRDG--TTASESEANNLPSPFEPLENITAKFISKGLEKKDVAVLSGAHTFGFAQ 743
Query: 260 C 260
C
Sbjct: 744 C 746
>TC17568 similar to UP|Q8RVP6 (Q8RVP6) Gaiacol peroxidase , partial (33%)
Length = 672
Score = 112 bits (279), Expect = 1e-25
Identities = 54/115 (46%), Positives = 81/115 (69%)
Frame = +2
Query: 79 TINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLD 138
T + +LR GFYS++CP AE I+ D + + P+++A+++R QFHDCFV GCDAS+LLD
Sbjct: 329 TSSSDLRPGFYSNTCPEAEYIVQDVMKKALFREPRSVASVMRFQFHDCFVNGCDASMLLD 508
Query: 139 STPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMA 193
TP+ EK + N L+ ++VD+IK LE++CPG+VSCAD ++ S +A+A
Sbjct: 509 DTPDMLG-EKLALSNINSLRSFEVVDEIKEALEKKCPGVVSCADIIIMASRDAVA 670
>TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
partial (47%)
Length = 604
Score = 108 bits (270), Expect = 2e-24
Identities = 57/129 (44%), Positives = 80/129 (61%)
Frame = +1
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L +GFY+ +CP AEKII D E + P A ++R+ FHDCFV GCDASILL+ST
Sbjct: 163 LEQGFYTKTCPKAEKIILDFVHEHIHNAPSLAAALIRMNFHDCFVRGCDASILLNST--S 336
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
E+ + N + ++G D +D IKS +E +CPG+VSCAD + + +++ G P K
Sbjct: 337 KQAERDAPPN-LTVRGFDFIDRIKSLVEAQCPGVVSCADIIALAARDSIVATGGPFWKVP 513
Query: 204 GGRRDSLYS 212
GRRD + S
Sbjct: 514 TGRRDGVIS 540
>TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%)
Length = 559
Score = 101 bits (251), Expect = 2e-22
Identities = 56/150 (37%), Positives = 81/150 (53%)
Frame = +3
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L E FY CP+ + V + ++LRL FHDCFV GCD S+LLD T +
Sbjct: 111 LSENFYVKKCPSVFNAVKSVVHSAVAKEARMGGSLLRLFFHDCFVNGCDGSVLLDDTSSF 290
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
EK++ N L+G D++D IKSK+E CPG+VSCAD + + +++A+ G P K
Sbjct: 291 KG-EKTAPPNSNSLRGFDVIDAIKSKVEAVCPGVVSCADVVAIAARDSVAILGGPYWKVK 467
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMV 233
GRRDS + + +P P S ++
Sbjct: 468 LGRRDSKTASFNAANSGVIPSPFSSLSDLI 557
>TC18385 similar to UP|Q9XFL6 (Q9XFL6) Peroxidase 5, partial (38%)
Length = 424
Score = 100 bits (250), Expect = 3e-22
Identities = 50/116 (43%), Positives = 75/116 (64%)
Frame = +3
Query: 81 NPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDST 140
N L FYS +CPN I+++A + ++++ + A+++RL FHDCFV G DASILLD
Sbjct: 72 NGQLNSTFYSSTCPNVSFIVSNAVQQALQSDSRIGASLIRLHFHDCFVNGGDASILLDQG 251
Query: 141 PNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAG 196
N EK++ N ++G D+VD IKS LE CPGIVSCAD + + +++++G
Sbjct: 252 GNITQSEKNAAPNFNSIRGFDVVDTIKSALENSCPGIVSCADILALAAESSVSMSG 419
>AV412471
Length = 430
Score = 99.0 bits (245), Expect = 1e-21
Identities = 47/114 (41%), Positives = 74/114 (64%)
Frame = +2
Query: 83 NLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPN 142
+L +Y SCP + I+ + + ++ +P A ++R+ FHDCF+ GCD S+LLDST +
Sbjct: 95 DLNMNYYLFSCPFVDPIVKNKVSTALQNDPTLAAGLIRMHFHDCFIEGCDGSVLLDSTKD 274
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAG 196
+ EK S N + L+G +++DDIK +LE+ CPG+VSCAD + S +A+ AG
Sbjct: 275 -NTAEKDSPAN-LSLRGYEIIDDIKEELEKRCPGVVSCADIVAMASRDAVFFAG 430
>AV426241
Length = 429
Score = 98.2 bits (243), Expect = 2e-21
Identities = 54/121 (44%), Positives = 77/121 (63%), Gaps = 1/121 (0%)
Frame = +3
Query: 87 GFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNV 146
GFYS SCP+ E I+ A V+T+ + A +LRL F DCFV GCDASIL+ G+
Sbjct: 81 GFYSKSCPSIESIVKSTVASHVKTDFEYAAGLLRLHFRDCFVRGCDASILI----AGNGT 248
Query: 147 EKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA-GMPRQKPLGG 205
EK + N LKG +++D+ K+KLE +CPG+VSCAD + + +++ L+ G+ Q P G
Sbjct: 249 EKQAPPNR-SLKGYEVIDEAKAKLEAQCPGVVSCADILALAARDSVVLSGGLSWQVPTGR 425
Query: 206 R 206
R
Sbjct: 426 R 428
>AV414604
Length = 427
Score = 97.4 bits (241), Expect = 3e-21
Identities = 49/112 (43%), Positives = 73/112 (64%)
Frame = +2
Query: 83 NLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPN 142
+LR+ FY SC AE+I+ + V + P+ A +LR+ FHDCFV GCD S+LL+ST
Sbjct: 83 SLRKQFYRKSCSQAEQIVKTTIQQHVSSRPELPAKLLRMHFHDCFVRGCDGSVLLNSTA- 259
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMAL 194
G+ EK + N + L G D++D+IK LE +CP IVSCAD + + +A+++
Sbjct: 260 GNTAEKDAIPN-LSLSGFDVIDEIKEALEAKCPKIVSCADILALAARDAVSV 412
>TC17595 similar to UP|Q9XFI7 (Q9XFI7) Peroxidase (Fragment), partial (42%)
Length = 657
Score = 97.1 bits (240), Expect = 5e-21
Identities = 51/130 (39%), Positives = 78/130 (59%), Gaps = 1/130 (0%)
Frame = +1
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L+ GFYS++CP+AE I+ V ++P A +LRL FHDCFV GCD SIL++ NG
Sbjct: 187 LQVGFYSNTCPHAESIVQAVVRGAVASDPNMAAVLLRLHFHDCFVEGCDGSILIE---NG 357
Query: 144 DNVEKSSF-FNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKP 202
D EK +F G+ +G ++++ K++ E CP +VSCAD + + +++ + P +
Sbjct: 358 DQSEKLAFGHQGV--RGFEVIERAKAQSEASCPDVVSCADIVALAATDSIVMGNGPEYQV 531
Query: 203 LGGRRDSLYS 212
GRRD S
Sbjct: 532 PTGRRDGSVS 561
>BP051104
Length = 566
Score = 93.6 bits (231), Expect = 5e-20
Identities = 54/145 (37%), Positives = 82/145 (56%)
Frame = +2
Query: 128 VVGCDASILLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFL 187
V GCDASIL D T N E+++ N +G +++D IK+ LE++CPG+VSCAD +
Sbjct: 71 VNGCDASILXDDTSNFIG-EQTAAANNRSARGFNVIDGIKANLEKQCPGVVSCADVLALA 247
Query: 188 SFEAMALAGMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMV 247
+ +++ G P + GRRDS + + +N +P P S ++ F +G S ++V
Sbjct: 248 ARDSVVQLGGPSWEVGLGRRDST-TASRGTANNTIPGPFLSLSGLITNFANQGLSVTDLV 424
Query: 248 ILLGAHSIGAAHCDIFMDRVYNFKN 272
L GAH+IG A C F +YN N
Sbjct: 425 ALSGAHTIGLAQCKNFRAHIYNDSN 499
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,519,890
Number of Sequences: 28460
Number of extensions: 92843
Number of successful extensions: 542
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 487
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 492
length of query: 388
length of database: 4,897,600
effective HSP length: 92
effective length of query: 296
effective length of database: 2,279,280
effective search space: 674666880
effective search space used: 674666880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0076a.9


