

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0074.3
(267 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
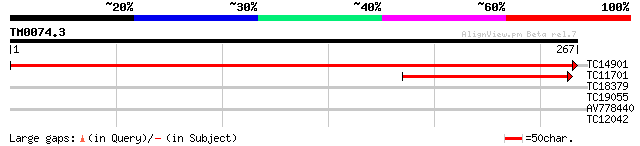
Score E
Sequences producing significant alignments: (bits) Value
TC14901 similar to UP|CAE47100 (CAE47100) 4,5-DOPA dioxygenase e... 553 e-159
TC11701 weakly similar to UP|CAE47100 (CAE47100) 4,5-DOPA dioxyg... 133 3e-32
TC18379 similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich glycop... 29 0.73
TC19055 28 1.2
AV778440 27 4.7
TC12042 similar to GB|CAB87983.1|7576235|ATH276134 Pex10p {Arabi... 26 8.0
>TC14901 similar to UP|CAE47100 (CAE47100) 4,5-DOPA dioxygenase extradiol,
partial (71%)
Length = 1276
Score = 553 bits (1426), Expect = e-159
Identities = 267/267 (100%), Positives = 267/267 (100%)
Frame = +1
Query: 1 MALKDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVV 60
MALKDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVV
Sbjct: 202 MALKDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVV 381
Query: 61 DSTNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWV 120
DSTNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWV
Sbjct: 382 DSTNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWV 561
Query: 121 PLLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERH 180
PLLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERH
Sbjct: 562 PLLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERH 741
Query: 181 ATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGEN 240
ATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGEN
Sbjct: 742 ATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGEN 921
Query: 241 SKAKLIHSSIDLGSLSYASYQFTSDVI 267
SKAKLIHSSIDLGSLSYASYQFTSDVI
Sbjct: 922 SKAKLIHSSIDLGSLSYASYQFTSDVI 1002
>TC11701 weakly similar to UP|CAE47100 (CAE47100) 4,5-DOPA dioxygenase
extradiol, partial (29%)
Length = 443
Score = 133 bits (335), Expect = 3e-32
Identities = 57/80 (71%), Positives = 71/80 (88%)
Frame = +3
Query: 186 PWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAKL 245
PWAV+F +WLK +LL GRYE+VN YE+KAP+AK AHPWPDHF+PLHVA+GAAGEN+KAK+
Sbjct: 21 PWAVDFMSWLKSSLLHGRYEEVNQYEEKAPYAKMAHPWPDHFFPLHVAMGAAGENAKAKV 200
Query: 246 IHSSIDLGSLSYASYQFTSD 265
+H S D GS+SYAS+ FT+D
Sbjct: 201 VHDSWDGGSISYASFGFTAD 260
>TC18379 similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich glycoprotein
DZ-HRGP precursor, partial (8%)
Length = 584
Score = 29.3 bits (64), Expect = 0.73
Identities = 18/58 (31%), Positives = 26/58 (44%)
Frame = +1
Query: 174 LRALERHATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLH 231
L LERH A ++F+ EA+ G Y + Y P+ + P+P YP H
Sbjct: 64 LSVLERHGKHAEIKYIKFNG---EAMDMGSYYNYGDYGYAPPYGMEFSPYPSLPYPQH 228
>TC19055
Length = 408
Score = 28.5 bits (62), Expect = 1.2
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = +1
Query: 4 KDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPR 39
+DT SHG P LS+ +S V + +W K + P+
Sbjct: 73 EDTSDCSHGXPGLSMTKSDVTAAEISTWAKMITKPK 180
>AV778440
Length = 489
Score = 26.6 bits (57), Expect = 4.7
Identities = 17/64 (26%), Positives = 29/64 (44%), Gaps = 2/64 (3%)
Frame = -1
Query: 71 YGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDK--KRGLDHGAWVPLLLMYPE 128
YG P P+ ++ + HLA K+ + EGG K +R + A +P + P
Sbjct: 366 YGQPTPVADVESASVHLKHLAAVAKDAVAEGGSIPGSRAKLWRRIREFNACIPYSGVLPN 187
Query: 129 ADIP 132
++P
Sbjct: 186 IEVP 175
>TC12042 similar to GB|CAB87983.1|7576235|ATH276134 Pex10p {Arabidopsis
thaliana;} , partial (19%)
Length = 582
Score = 25.8 bits (55), Expect = 8.0
Identities = 15/41 (36%), Positives = 19/41 (45%), Gaps = 3/41 (7%)
Frame = +1
Query: 145 THHYNIGKALAPLKDEGVLI---VGSGSAVHNLRALERHAT 182
THH + G L L +EG L GS + + E HAT
Sbjct: 10 THHTSAGHGLPVLNEEGTLASPDTDKGSWISESSSSEYHAT 132
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,139,738
Number of Sequences: 28460
Number of extensions: 75203
Number of successful extensions: 304
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 304
length of query: 267
length of database: 4,897,600
effective HSP length: 89
effective length of query: 178
effective length of database: 2,364,660
effective search space: 420909480
effective search space used: 420909480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0074.3


