

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0073.15
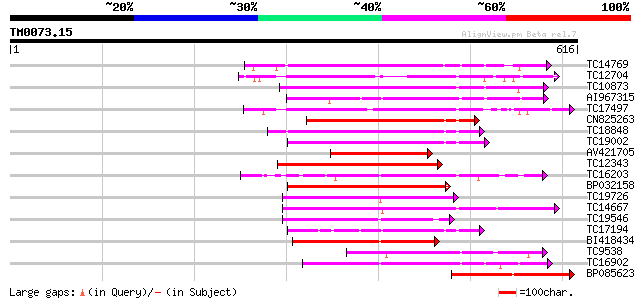
(616 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 168 3e-42
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 164 5e-41
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 162 1e-40
AI967315 161 3e-40
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 160 6e-40
CN825263 158 2e-39
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 158 3e-39
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 153 9e-38
AV421705 151 3e-37
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 146 1e-35
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 145 2e-35
BP032158 144 3e-35
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 144 5e-35
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 142 2e-34
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 142 2e-34
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 141 3e-34
BI418434 138 3e-33
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 132 2e-31
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 129 1e-30
BP085623 127 7e-30
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 168 bits (425), Expect = 3e-42
Identities = 119/360 (33%), Positives = 187/360 (51%), Gaps = 27/360 (7%)
Frame = +3
Query: 256 GALLICIT-----ICIFRRKLSPIVSKHWKAKKVDQDI--------EAFIRNNGPQAIKR 302
G+LLI + +C +R+KL P K ++ +I + FI++ QA
Sbjct: 1962 GSLLITLAFGVLFVCRYRQKLIPWEGFAGKKYPMETNIIFSLPSKDDFFIKSVSIQA--- 2132
Query: 303 YSYSEIKKVTNSFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIG 361
++ I+ T +K+ +G+GG+G VY+G LN+ VAVKV +++ G +EF NE+ +
Sbjct: 2133 FTLEYIEVATERYKTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELNLLS 2312
Query: 362 RTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETN-LSWERLHKIAEGIAK 420
H N+V LLG+C E ++ L+Y FM NGSL+ + + + L W IA G A+
Sbjct: 2313 AIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAAR 2492
Query: 421 GLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLC-SETHSIISMHDARGTIG 479
GL YLH ++H DIK SNILLD + C K+ADFG +K E S +S+ + RGT G
Sbjct: 2493 GLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSL-EVRGTAG 2669
Query: 480 YIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVE 539
Y+ PE + +S KSDV+S+G+++LEIV ++ + N +E W +I
Sbjct: 2670 YLDPEYYKTQ--QLSEKSDVFSFGVVLLEIVSGREPL-NIKRPRTEWSLVEWATPYIR-- 2834
Query: 540 SNLAWHDGMSIEE-----------NEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSI 588
G ++E E ++V V L C++ + RP M +V LE ++
Sbjct: 2835 -------GSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELEDAL 2993
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 164 bits (414), Expect = 5e-41
Identities = 136/396 (34%), Positives = 187/396 (46%), Gaps = 47/396 (11%)
Frame = +1
Query: 249 GSIAGGIGALLICITI-------CIFRRK--------LSPIVSKHWKAKKVDQDIEAFIR 293
GS+AG I A +ICITI CI R+K + + HWK K+ D+DI+
Sbjct: 1240 GSLAGSI-AFIICITILGLATVTCIRRKKNEREDEGGIETRIINHWKDKRGDEDIDL--- 1407
Query: 294 NNGPQAIKRYSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG- 350
+ +S I TN F +KLG+GG+G VYKG L N +AVK L+ + G G
Sbjct: 1408 ------ATIFDFSTISSTTNHFSESNKLGEGGFGPVYKGVLANGQEIAVKRLSNTSGQGM 1569
Query: 351 QEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWER 410
+EF NEV I R H N+V LLG + + LIYEFM N SL+ F F++ L
Sbjct: 1570 EEFKNEVKLIARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFI----FDSRL---- 1725
Query: 411 LHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIIS 470
RI+H D+K SNILLD PKI+DFGLA++ +
Sbjct: 1726 ---------------------RIIHRDLKTSNILLDSEMNPKISDFGLARIFTGDQVEAK 1842
Query: 471 MHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQ--------NIRNEASD 522
GT GY++PE G S KSDV+S+G+++LEI+ K+ + RN S
Sbjct: 1843 TKRVMGTYGYMSPEY--AVHGSFSVKSDVFSFGVIVLEIISGKKIGRFCDPHHHRNLLSH 2016
Query: 523 SSETYFPHWIYKHI------EVESNLAWH---------------DGMSIEENEICKKMVI 561
SS F ++ K + V++ AW DG++I + + +
Sbjct: 2017 SSN--FAVFLIKALRICMFENVKNRKAWRLWIEERPLELVDELLDGLAIPTEIL--RYIH 2184
Query: 562 VGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKP 597
+ L C+Q P RP M VV ML G E PKP
Sbjct: 2185 IALLCVQQRPEYRPDMLSVVLMLNGEKEL----PKP 2280
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 162 bits (411), Expect = 1e-40
Identities = 99/299 (33%), Positives = 159/299 (53%), Gaps = 7/299 (2%)
Frame = +1
Query: 294 NNGPQAIKRYSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG- 350
+NG A + + E+ T +FK +G+GG+G+VYKG L VAVK L+ G
Sbjct: 388 SNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGF 567
Query: 351 QEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETN-LSWE 409
QEF+ EV+ + H N+V L+G+C +G ++ L+YE+MP GSLE + + + L+W
Sbjct: 568 QEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWS 747
Query: 410 RLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSII 469
K+A G A+GLEYLH + +++ D+K +NILLD F PK++DFGLAKL +
Sbjct: 748 TRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTH 927
Query: 470 SMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFP 529
GT GY APE G ++ KSD+YS+G+++LE++ ++ I + + E
Sbjct: 928 VSTRVMGTYGYCAPEY--AMSGKLTLKSDIYSFGVVLLELLTGRRAI-DTSRRPGEQNLV 1098
Query: 530 HWIYKHIEVESNLAWHDGMSIE---ENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
W + ++ + + + + C+Q P RP ++ +V LE
Sbjct: 1099SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 1275
>AI967315
Length = 1308
Score = 161 bits (408), Expect = 3e-40
Identities = 102/290 (35%), Positives = 160/290 (55%), Gaps = 5/290 (1%)
Frame = +1
Query: 301 KRYSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLNAS---KGNGQEFLN 355
K +SY E+ TN F S+ +G+GGY +VYKG L + +AVK L + + +EFL
Sbjct: 106 KCFSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLT 285
Query: 356 EVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIA 415
E+ +IG H NV+ LLG C++ L++E GS+ H + L W+ +KI
Sbjct: 286 EIGTIGHVCHSNVMPLLGCCIDNGLY-LVFELSTVGSVASLIHDEKMAP-LDWKTRYKIV 459
Query: 416 EGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDAR 475
G A+GL YLHKGC RI+H DIK SNILL ++F P+I+DFGLAK + S+
Sbjct: 460 LGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIE 639
Query: 476 GTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKH 535
GT G++APE + G V K+DV+++G+ +LE++ ++ + + S S + I
Sbjct: 640 GTFGHLAPEYYMH--GVVDEKTDVFAFGVFLLEVISGRKPV--DGSHQSLHTWAKPILSK 807
Query: 536 IEVESNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
E+E + + + ++ CI+ + RP MS+V+E++E
Sbjct: 808 WEIEKLVDPRLEGCYDVTQF-NRVAFAASLCIRASSTWRPTMSEVLEVME 954
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 160 bits (405), Expect = 6e-40
Identities = 128/383 (33%), Positives = 183/383 (47%), Gaps = 24/383 (6%)
Frame = +1
Query: 255 IGALLICITICIFRRKLSPI------VSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYSEI 308
I L + I+ CI R+K + HWK K+ D+DI+ + +S I
Sbjct: 1501 ITILGLAISTCIQRKKNKRGDEGEIGIINHWKDKRGDEDIDL---------ATIFDFSTI 1653
Query: 309 KKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRTSH 365
TN F +KLG+GG+G VYKG L N +AVK L+ + G G +EF NE+ I R H
Sbjct: 1654 SSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNEIKLIARLQH 1833
Query: 366 VNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYL 425
N+V L G + + + + M K + W + +I +GIA+GL YL
Sbjct: 1834 RNLVKLFGCSVHQDENSHANKKM------KILLDSTRSKLVDWNKRLQIIDGIARGLLYL 1995
Query: 426 HKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEV 485
H+ RI+H D+K SNILLD PKI+DFGLA++ GT GY+ PE
Sbjct: 1996 HQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEY 2175
Query: 486 WNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAWH 545
G S KSDV+S+G+++LEI+ K+ Y PH H+ + S+ AW
Sbjct: 2176 --AVHGSFSIKSDVFSFGVIVLEIISGKK--------IGRFYDPH---HHLNLLSH-AWR 2313
Query: 546 DGMSIEE------NEICKKMVI---------VGLWCIQTIPSNRPPMSKVVEMLEGSIEQ 590
+ IEE +E+ VI V L C+Q P NRP M +V ML G ++
Sbjct: 2314 --LWIEERPLELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLNGE-KE 2484
Query: 591 LQIPPKPFMFSPTKTEVESGTTS 613
L P P ++ + G+ S
Sbjct: 2485 LPKPRLPAFYTGKHDPIWLGSPS 2553
>CN825263
Length = 663
Score = 158 bits (400), Expect = 2e-39
Identities = 91/191 (47%), Positives = 121/191 (62%), Gaps = 3/191 (1%)
Frame = +1
Query: 323 GYGQVYKGNLNNNCPVAVKVLNAS-KGNGQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKK 381
G+G VYKG LN+ VAVK+L + G+EFL EV + R H N+V L+G C+E Q +
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 382 ALIYEFMPNGSLEKFTHKKNFETN-LSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKP 440
LIYE +PNGS+E H + ET L W KIA G A+GL YLH+ N ++H D K
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 441 SNILLDKNFCPKIADFGLAKLC-SETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDV 499
SNILL+ +F PK++DFGLA+ E + IS H GT GY+APE G + KSDV
Sbjct: 361 SNILLECDFTPKVSDFGLARTALDEGNKHISTH-VMGTFGYLAPEY--AMTGHLLVKSDV 531
Query: 500 YSYGMLILEIV 510
YSYG+++LE++
Sbjct: 532 YSYGVVLLELL 564
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 158 bits (399), Expect = 3e-39
Identities = 93/240 (38%), Positives = 142/240 (58%), Gaps = 4/240 (1%)
Frame = +2
Query: 281 AKKVDQDIEAFIRNNGPQAIKRYSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPV 338
++KV++ +F N I ++Y E+ T F +KLG+GG+G VY G ++ +
Sbjct: 206 SEKVEEGPTSFGSVNNSWRI--FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQI 379
Query: 339 AVKVLNASKGNGQ-EFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFT 397
AVK L A + EF EV +GR H N++ L G+C+ ++ ++Y++MPN SL
Sbjct: 380 AVKKLKAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHL 559
Query: 398 HKK-NFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADF 456
H + E L+W++ KIA G A+G+ YLH I+H DIK SN+LL+ +F P +ADF
Sbjct: 560 HGQFAVEVQLNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADF 739
Query: 457 GLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNI 516
G AKL E S ++ +GT+GY+APE +G VS DVYS+G+L+LE+V ++ I
Sbjct: 740 GFAKLIPEGVSHMTTR-VKGTLGYLAPEY--AMWGKVSESCDVYSFGILLLELVTGRKPI 910
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 153 bits (386), Expect = 9e-38
Identities = 87/223 (39%), Positives = 131/223 (58%), Gaps = 4/223 (1%)
Frame = +1
Query: 303 YSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNA-SKGNGQEFLNEVVS 359
+S E+ TN+F +KLG+GG+G VY G L + +AVK L S EF EV
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEI 207
Query: 360 IGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTH-KKNFETNLSWERLHKIAEGI 418
+ R H N+++L G+C EGQ++ ++Y++MPN SL H + + E L W R IA G
Sbjct: 208 LARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGS 387
Query: 419 AKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTI 478
A+G+ YLH I+H DIK SN+LLD +F ++ADFG AKL + + ++ +GT+
Sbjct: 388 AEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTR-VKGTL 564
Query: 479 GYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEAS 521
GY+APE G + DV+S+G+L+LE+ K+ + +S
Sbjct: 565 GYLAPEY--AMLGKANECCDVFSFGILLLELASGKKPLEKLSS 687
>AV421705
Length = 341
Score = 151 bits (381), Expect = 3e-37
Identities = 65/111 (58%), Positives = 94/111 (84%)
Frame = +3
Query: 349 NGQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSW 408
NGQ+F++EV +IGR H+NVV L+G+C++G+K+AL+YEFMPNGSL+K+ K LS+
Sbjct: 9 NGQDFISEVATIGRIHHINVVRLVGYCVDGKKRALVYEFMPNGSLDKYIFSKEGSAWLSY 188
Query: 409 ERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLA 459
+++++I+ GIA+G+ YLH+GC+ +ILHFDIKP NILLDK+F PK++DFGLA
Sbjct: 189 DQIYEISLGIARGVAYLHQGCDMQILHFDIKPHNILLDKDFIPKVSDFGLA 341
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 146 bits (368), Expect = 1e-35
Identities = 74/182 (40%), Positives = 113/182 (61%), Gaps = 3/182 (1%)
Frame = +3
Query: 292 IRNNGPQAIKRYSYSEIKKVTNSFKS--KLGQGGYGQVYKGNLNNNCPVAVKVLNASKGN 349
I+N + K +SY + T +F + KLG+GG+G V+KG LN+ +AVK L+
Sbjct: 174 IQNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQ 353
Query: 350 GQ-EFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSW 408
G+ +F+NE + R H NVV+L G+C G +K L+YE++P SL+K + + L W
Sbjct: 354 GRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQLDW 533
Query: 409 ERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSI 468
+R I G+A+GL YLH+ + I+H DIK +NILLD+ + PKIADFGLA++ E +
Sbjct: 534 KRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARIFPEDQTH 713
Query: 469 IS 470
++
Sbjct: 714 VN 719
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 145 bits (365), Expect = 2e-35
Identities = 115/353 (32%), Positives = 174/353 (48%), Gaps = 19/353 (5%)
Frame = +2
Query: 251 IAGGIGALLICITICIFRRKLSPIVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYSEIKK 310
IA LL+ +T+ + R++ ++ WK + AF R IK E K
Sbjct: 2054 IALATAVLLVAVTVHVVRKRRLHR-AQAWK-------LTAFQRLE----IKAEDVVECLK 2197
Query: 311 VTNSFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQE---FLNEVVSIGRTSHVN 367
N +G+GG G VY+G++ N VA+K L +G+G+ F E+ ++G+ H N
Sbjct: 2198 EENI----IGKGGAGIVYRGSMPNGTDVAIKRL-VGQGSGRNDYGFRAEIETLGKIRHRN 2362
Query: 368 VVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHK 427
++ LLG+ L+YE+MPNGSL ++ H +L WE +KIA A+GL Y+H
Sbjct: 2363 IMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAK-GGHLRWEMRYKIAVEAARGLCYMHH 2539
Query: 428 GCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWN 487
C+ I+H D+K +NILLD +F +ADFGLAK + + SM G+ GYIAPE
Sbjct: 2540 DCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEY-- 2713
Query: 488 RNFGGVSHKSDVYSYGMLILE----------------IVGAKQNIRNEASDSSETYFPHW 531
V KSDVYS+G+++LE IVG +E S S+T
Sbjct: 2714 AYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLA 2893
Query: 532 IYKHIEVESNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEML 584
+ V+ L+ + S+ M + + C++ + RP M +VV ML
Sbjct: 2894 V-----VDPRLSGYPLTSVIH------MFNIAMMCVKEMGPARPTMREVVHML 3019
>BP032158
Length = 555
Score = 144 bits (364), Expect = 3e-35
Identities = 83/181 (45%), Positives = 115/181 (62%), Gaps = 4/181 (2%)
Frame = +2
Query: 303 YSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNA-SKGNGQEFLNEVVS 359
+S +IK TN+F +K+G+GG+G VYKG L+ +AVK L++ SK +EF+NE+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFINEIGM 193
Query: 360 IGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTH-KKNFETNLSWERLHKIAEGI 418
I H N+V L G C+EG + L+YE+M N SL + + + NL+W KI GI
Sbjct: 194 ISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVGI 373
Query: 419 AKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTI 478
AKGL YLH+ +I+H DIK +N+LLDK+ KI+DFGLAKL E ++ IS A GTI
Sbjct: 374 AKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIA-GTI 550
Query: 479 G 479
G
Sbjct: 551 G 553
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 144 bits (362), Expect = 5e-35
Identities = 82/199 (41%), Positives = 118/199 (59%), Gaps = 8/199 (4%)
Frame = +3
Query: 297 PQAIKRYSYSEIKKVTNSF--KSKLGQGGYGQVYKGNL-NNNCPVAVKVLNASKGNG-QE 352
P + ++Y E+KK TN+F K KLGQGGYG VY+G L VAVK+ + K +
Sbjct: 114 PGTPREFNYVELKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDD 293
Query: 353 FLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKN---FETNLSWE 409
FL+E++ I R H ++V L G+C + L+Y++MPNGSL+ + T LSW
Sbjct: 294 FLSELIIINRLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWP 473
Query: 410 RLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAK-LCSETHSI 468
+KI G+A L YLH + +++H D+K SNI+LD F K+ DFGLA+ L +E S
Sbjct: 474 LRYKIISGVASALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISY 653
Query: 469 ISMHDARGTIGYIAPEVWN 487
+ GT+GYIAPE ++
Sbjct: 654 TELEGVHGTMGYIAPECFH 710
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 142 bits (357), Expect = 2e-34
Identities = 100/313 (31%), Positives = 156/313 (48%), Gaps = 12/313 (3%)
Frame = +2
Query: 297 PQAIKRYSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLNASKG--NGQE 352
P + S E+K+ T++F SK +G+G YG+VY LN+ VAVK L+ S E
Sbjct: 305 PIEVPALSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNE 484
Query: 353 FLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTH-KKNFE-----TNL 406
FL +V + R + N V L G+C+EG + L YEF GSL H +K + L
Sbjct: 485 FLTQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTL 664
Query: 407 SWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETH 466
W + +IA A+GLEYLH+ I+H DI+ SN+L+ +++ KIADF L+ +
Sbjct: 665 DWIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMA 844
Query: 467 SIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSET 526
+ + GT GY APE G ++ KSDVYS+G+++LE++ ++ + + ++
Sbjct: 845 ARLHSTRVLGTFGYHAPEY--AMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQS 1018
Query: 527 YFPHWIYKHIEVESNLAWHDGMSIEE--NEICKKMVIVGLWCIQTIPSNRPPMSKVVEML 584
W + + D E + K+ V C+Q RP MS VV+ L
Sbjct: 1019LVT-WATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKAL 1195
Query: 585 EGSIEQLQIPPKP 597
+ ++ P P
Sbjct: 1196QPLLKTPAAAPAP 1234
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 142 bits (357), Expect = 2e-34
Identities = 79/190 (41%), Positives = 115/190 (59%), Gaps = 3/190 (1%)
Frame = +1
Query: 297 PQAIKRYSYSEIKKVTNSFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQ-EFLN 355
P I +YSY +I+K T +F + LGQG +G VYK + VAVKVL + G+ EF
Sbjct: 181 PSGIPKYSYKDIQKATQNFTTILGQGSFGTVYKATMPTGEVVAVKVLAPNSKQGEHEFQT 360
Query: 356 EVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIA 415
EV +GR H N+VNL+GFC++ ++ L+Y+FM NGSL + + E LSW+ +IA
Sbjct: 361 EVHLLGRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGE--EKELSWDERLQIA 534
Query: 416 EGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDA- 474
I+ G+EYLH+G ++H D+K +NILLD + +ADFGL+K I ++
Sbjct: 535 MDISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSK-----EEIFDGRNSG 699
Query: 475 -RGTIGYIAP 483
+GT GY+ P
Sbjct: 700 LKGTYGYMDP 729
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 141 bits (356), Expect = 3e-34
Identities = 82/216 (37%), Positives = 125/216 (56%), Gaps = 2/216 (0%)
Frame = +1
Query: 303 YSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQEFLNEVVSI 360
+SY E+ K TN+F +K+GQGG+G VY L A+K ++ EFL E+ +
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAVYYAELRGK-KTAIKKMDVQAST--EFLCELKVL 1221
Query: 361 GRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAK 420
H+N+V L+G+C+EG L+YE + NG+L ++ H E L W +IA A+
Sbjct: 1222 THVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLHGSGKEP-LPWSSRVQIALDAAR 1395
Query: 421 GLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGY 480
GLEY+H+ +H D+K +NIL+DKN K+ADFGL KL +S + GT GY
Sbjct: 1396 GLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTR-LVGTFGY 1572
Query: 481 IAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNI 516
+ PE +G +S K DVY++G+++ E++ AK +
Sbjct: 1573 MPPEY--AQYGDISPKIDVYAFGVVLFELISAKNAV 1674
>BI418434
Length = 478
Score = 138 bits (347), Expect = 3e-33
Identities = 72/160 (45%), Positives = 97/160 (60%)
Frame = +2
Query: 308 IKKVTNSFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQEFLNEVVSIGRTSHVN 367
++ T F K+G GG+G V++G L+++ VAVK L G +EF EV +IG HVN
Sbjct: 2 LQLATRGFSEKVGHGGFGTVFQGELSDSSLVAVKRLERPGGGEKEFRAEVSTIGNIQHVN 181
Query: 368 VVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHK 427
+V L GFC E + L+YE+M NG+L + K LSW+ ++A G AKG+ YLH+
Sbjct: 182 LVRLRGFCSENSHRLLVYEYMHNGALSAYLRKDG--PCLSWDVRFQVAIGTAKGIAYLHE 355
Query: 428 GCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHS 467
ILH DIKP NILLD +F K++DFGLAKL S
Sbjct: 356 ELRCCILHCDIKPENILLDSDFTAKVSDFGLAKLIGRDFS 475
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 132 bits (331), Expect = 2e-31
Identities = 83/232 (35%), Positives = 125/232 (53%), Gaps = 14/232 (6%)
Frame = +2
Query: 367 NVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLS----------WERLHKIAE 416
N+V LL L+YE++ N SL+K+ H K +++S W + KIA
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAI 184
Query: 417 GIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARG 476
G A+GL Y+H C+ I+H D+K SNILLDK F K+ADFGLA++ + + M G
Sbjct: 185 GAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIG 364
Query: 477 TIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEI-VGAKQNIRNEASDSSETYFPHWIYKH 535
T GYIAPE +S K DVYS+G+++LE+ G + N ++ S +E W ++H
Sbjct: 365 TFGYIAPEYVQTT--RISEKVDVYSFGVVLLELTTGKEANYGDQHSSLAE-----WAWRH 523
Query: 536 IEVESNLAWHDGMSIEENEICKKMVIV---GLWCIQTIPSNRPPMSKVVEML 584
I + SN+ + E +M V G+ C T+P+ RP M +V+ +L
Sbjct: 524 ILIGSNVXDLLXKDVMEASYIDEMCSVFKLGVMCTATLPATRPSMKEVLPIL 679
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 129 bits (325), Expect = 1e-30
Identities = 85/275 (30%), Positives = 142/275 (50%), Gaps = 4/275 (1%)
Frame = +3
Query: 319 LGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRTSHVNVVNLLGFCLE 377
LG GG+G+VY G ++ VA+K N G EF E+ + + H ++V+L+G+C E
Sbjct: 15 LGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCEE 194
Query: 378 GQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFD 437
+ L+Y+ M G+L + +K + L W++ +I G A+GL YLH G I+H D
Sbjct: 195 NTEMILVYDHMAYGTLREHLYKTQ-KPPLPWKQRLEICIGAARGLHYLHTGAKYTIIHRD 371
Query: 438 IKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKS 497
+K +NILLD+ + K++DFGL+K + +G+ GY+ PE + R ++ KS
Sbjct: 372 VKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQ--QLTDKS 545
Query: 498 DVYSYGMLILEIVGAKQNIRNEASDSSETYFPHW---IYKHIEVESNLAWHDGMSIEENE 554
DVYS+G+++ EI+ A+ + N + + W Y ++ L + I E
Sbjct: 546 DVYSFGVVLFEILCARPAL-NPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIAP-E 719
Query: 555 ICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIE 589
KK + C+ RP M V+ LE +++
Sbjct: 720 CFKKFAETAMKCVSDQGIERPSMGDVLWNLEFALQ 824
>BP085623
Length = 528
Score = 127 bits (318), Expect = 7e-30
Identities = 58/133 (43%), Positives = 89/133 (66%)
Frame = -3
Query: 481 IAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVES 540
+APE++ +N GGVS+K DVYS+GML++E+ ++N+ SS+ YFP WIY + ++
Sbjct: 526 MAPELFYKNIGGVSYKXDVYSFGMLLMEMASNRKNLNPHVEHSSQLYFPFWIYDQLGKQN 347
Query: 541 NLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKPFMF 600
++ D + EE +I KKM++V LWCIQ P +RP M +VVEMLEG +E L++PPKP ++
Sbjct: 346 DIELED-VKEEEVKIAKKMIMVALWCIQLKPDDRPSMKEVVEMLEGDVENLEMPPKPSLY 170
Query: 601 SPTKTEVESGTTS 613
+ G+ S
Sbjct: 169 PHETIAGDQGSNS 131
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,926,378
Number of Sequences: 28460
Number of extensions: 215436
Number of successful extensions: 1565
Number of sequences better than 10.0: 299
Number of HSP's better than 10.0 without gapping: 1364
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1377
length of query: 616
length of database: 4,897,600
effective HSP length: 96
effective length of query: 520
effective length of database: 2,165,440
effective search space: 1126028800
effective search space used: 1126028800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0073.15


