

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
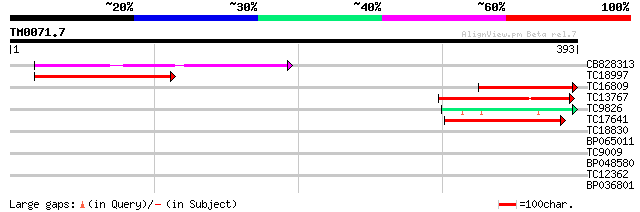
Query= TM0071.7
(393 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB828313 140 4e-34
TC18997 similar to GB|AAP31966.1|30387601|BT006622 At3g19640 {Ar... 115 1e-26
TC16809 weakly similar to UP|Q8H1G8 (Q8H1G8) MRS2-7, partial (18%) 97 5e-21
TC13767 similar to GB|AAN73219.1|25360983|AY150294 MRS2-10 {Arab... 86 8e-18
TC9826 similar to GB|AAN73212.1|25360813|AY150287 MRS2-2 {Arabid... 80 8e-16
TC17641 homologue to GB|AAP31966.1|30387601|BT006622 At3g19640 {... 69 2e-12
TC18830 weakly similar to UP|O40637 (O40637) Protease, partial (5%) 28 2.0
BP065011 28 2.6
TC9009 similar to PIR|T06773|T06773 ferredoxin-NADP reductase -... 27 4.5
BP048580 27 7.7
TC12362 weakly similar to UP|UBF1_HUMAN (P17480) Nucleolar trans... 27 7.7
BP036801 27 7.7
>CB828313
Length = 541
Score = 140 bits (353), Expect = 4e-34
Identities = 84/179 (46%), Positives = 106/179 (58%)
Frame = +2
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
V+KK R+W+L A+G +++ K+AIM R + ARDLRILDPLLSYPST
Sbjct: 44 VRKKATGVRAWLLLQANGDTEVVEAGKHAIMRRTGLPARDLRILDPLLSYPST------- 202
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRLPKVSSLHQQQGDGQEYVSSQHD 137
V+NLEHIKAII A EVLL + D +V P V+EL+ R+ L + +
Sbjct: 203 -VINLEHIKAIIIAHEVLLLNSRDPSVTPFVDELRARI-----LRHRHATTSNPKLEMDN 364
Query: 138 TEAAEEDESPFEFRALEVALEAICSFLAARTTELEMAAYPALDELTSKISSRNLDRVRK 196
E PFEF ALE LEA CS L LE A+PALD+LTSKIS+ NL+RVR+
Sbjct: 365 PEDGGMKILPFEFVALEACLEAACSVLENEAKTLEQEAHPALDKLTSKISTLNLERVRQ 541
>TC18997 similar to GB|AAP31966.1|30387601|BT006622 At3g19640 {Arabidopsis
thaliana;}, partial (17%)
Length = 406
Score = 115 bits (289), Expect = 1e-26
Identities = 57/98 (58%), Positives = 74/98 (75%)
Frame = +2
Query: 18 VKKKTQSSRSWILFDADGQGSLLDVDKYAIMHRVQIHARDLRILDPLLSYPSTILGREKA 77
V+KK R+W+L A+G +++ K+AIM R + ARDLRILDPLLSYPST+LGRE+A
Sbjct: 29 VRKKATGVRAWLLLQANGDTEVVEAGKHAIMRRTGLPARDLRILDPLLSYPSTVLGRERA 208
Query: 78 IVLNLEHIKAIITAEEVLLRDPTDENVVPVVEELQRRL 115
IV+NLEHIKAII A EVLL + D +V P V+EL+ R+
Sbjct: 209 IVINLEHIKAIIIAHEVLLLNSRDPSVTPFVDELRARI 322
>TC16809 weakly similar to UP|Q8H1G8 (Q8H1G8) MRS2-7, partial (18%)
Length = 490
Score = 97.1 bits (240), Expect = 5e-21
Identities = 46/68 (67%), Positives = 55/68 (80%)
Frame = +1
Query: 326 LELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKWVVIVSGVFSAIMFLLIIVYA 385
LELFL SGTV LS SLVAAIFGMNIPY+W + HG++FKWVVI +G+ A +FL I YA
Sbjct: 88 LELFLRSGTVSLSICSLVAAIFGMNIPYTWREGHGYIFKWVVIFAGMVCASLFLSIAAYA 267
Query: 386 RKKGLVGS 393
R++GLVGS
Sbjct: 268 RRQGLVGS 291
>TC13767 similar to GB|AAN73219.1|25360983|AY150294 MRS2-10 {Arabidopsis
thaliana;}, partial (21%)
Length = 652
Score = 86.3 bits (212), Expect = 8e-18
Identities = 44/94 (46%), Positives = 63/94 (66%)
Frame = +2
Query: 298 LTTLREYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYSWND 357
LT+L+EYIDDTED+INIQLDN RNQLIQ EL L++ T ++ + +VA IFGMN D
Sbjct: 2 LTSLKEYIDDTEDFINIQLDNVRNQLIQFELLLTTATFVVAIFGVVAGIFGMNFEIPLFD 181
Query: 358 NHGFMFKWVVIVSGVFSAIMFLLIIVYARKKGLV 391
F+WV+I++GV +F + + + + L+
Sbjct: 182 VPS-AFQWVLIITGVCGVFIFSAFVWFFKYRRLM 280
>TC9826 similar to GB|AAN73212.1|25360813|AY150287 MRS2-2 {Arabidopsis
thaliana;}, partial (13%)
Length = 1285
Score = 79.7 bits (195), Expect = 8e-16
Identities = 63/195 (32%), Positives = 77/195 (39%), Gaps = 101/195 (51%)
Frame = +3
Query: 300 TLREYIDDTEDYI----NIQLDNHRNQLIQ------------------------------ 325
T+ I++T Y+ ++QLDNHRNQLIQ
Sbjct: 75 TIYSNINNTWCYVAYA*HVQLDNHRNQLIQV*AFGPSLMRFFMPNPIHTLSEALICLQNH 254
Query: 326 --------------------LELFLSSGTVCLSFYSLVAAIFGMNIPYSWNDNHGFMFKW 365
LELFLSSGTV LS SLVAAIFGMNIPY+W + HG++FKW
Sbjct: 255 IL*CL*VLHLMFILVLPK**LELFLSSGTVSLSICSLVAAIFGMNIPYTWREGHGYIFKW 434
Query: 366 -----------------------------------------------VVIVSGVFSAIMF 378
VVI +G+ A +F
Sbjct: 435 VCMLRKKYSHFD*HVLPYTCVCV*EKESMSFFFLLNFVFSN*K**I*VVIFAGMVCASLF 614
Query: 379 LLIIVYARKKGLVGS 393
L I YAR+KGLVGS
Sbjct: 615 LSIAAYARRKGLVGS 659
>TC17641 homologue to GB|AAP31966.1|30387601|BT006622 At3g19640 {Arabidopsis
thaliana;}, partial (11%)
Length = 521
Score = 68.6 bits (166), Expect = 2e-12
Identities = 35/85 (41%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Frame = +2
Query: 302 REYIDDTEDYINIQLDNHRNQLIQLELFLSSGTVCLSFYSLVAAIFGMNIPYS-WNDNHG 360
REY+DDTEDYINI LD+ +N L+Q+ + L++ T+ +S + +VA IFGMNI +N +
Sbjct: 2 REYVDDTEDYINIMLDDKQNHLLQMGVMLTTATLVVSAFVVVAGIFGMNIKIELFNPDIA 181
Query: 361 FMFKWVVIVSGVFSAIMFLLIIVYA 385
M +++ V G + +FL ++ A
Sbjct: 182 GMREFLWTVGGSTAGTIFLYVVAIA 256
>TC18830 weakly similar to UP|O40637 (O40637) Protease, partial (5%)
Length = 850
Score = 28.5 bits (62), Expect = 2.0
Identities = 25/102 (24%), Positives = 46/102 (44%), Gaps = 1/102 (0%)
Frame = +2
Query: 126 GDGQEYVSSQHDTEAAEEDESPFEFRALEVAL-EAICSFLAARTTELEMAAYPALDELTS 184
G+G E + ++H TE E + L E + L+ TE + ++D +S
Sbjct: 38 GNGSEALINEHLTEGVEASYGNENIGEDDDLLPENSVTNLSWGATEEK-----SVDNQSS 202
Query: 185 KISSRNLDRVRKLKSAMTRLTARVQKVRDELEQLLDDDDDMA 226
+ ++ +R L + L VQK R+ +++ DDD+A
Sbjct: 203 NMEDPLIESIRSLLAVQEALEEEVQKFRETGIEVVSPDDDLA 328
>BP065011
Length = 527
Score = 28.1 bits (61), Expect = 2.6
Identities = 19/56 (33%), Positives = 28/56 (49%), Gaps = 7/56 (12%)
Frame = +3
Query: 231 SRKAGSSSPVSGSGAANWFGGSPTIGSKIS-------RASRASLATVRFDENDVEE 279
S+K SSSP G G GG+ T+ K++ RAS S V+ E +V++
Sbjct: 135 SKKRYSSSPRGGGGGGLRDGGAATVKVKVTQLV*GIRRASSPSSVEVKKTEEEVKQ 302
>TC9009 similar to PIR|T06773|T06773 ferredoxin-NADP reductase - garden
pea (fragment) {Pisum sativum;} ,
partial (33%)
Length = 585
Score = 27.3 bits (59), Expect = 4.5
Identities = 15/38 (39%), Positives = 22/38 (57%)
Frame = +2
Query: 230 LSRKAGSSSPVSGSGAANWFGGSPTIGSKISRASRASL 267
L +K+ PVS S ++W S + +I R+SRASL
Sbjct: 317 LCKKSSGDMPVSASATSDW-AQSYCLPIRIGRSSRASL 427
>BP048580
Length = 533
Score = 26.6 bits (57), Expect = 7.7
Identities = 8/29 (27%), Positives = 17/29 (58%)
Frame = -2
Query: 339 FYSLVAAIFGMNIPYSWNDNHGFMFKWVV 367
+ +L +G+++PY N+ F F W++
Sbjct: 157 YNALFLTFYGLSLPYICMANYSFWFMWMI 71
>TC12362 weakly similar to UP|UBF1_HUMAN (P17480) Nucleolar transcription
factor 1 (Upstream binding factor 1) (UBF-1)
(Autoantigen NOR-90), partial (3%)
Length = 493
Score = 26.6 bits (57), Expect = 7.7
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +3
Query: 216 EQLLDDDDDMADLYLSRKAGSSS 238
E DD+DDM + YL+ K+ SS+
Sbjct: 114 EDFEDDEDDMLEHYLAEKSDSST 182
>BP036801
Length = 540
Score = 26.6 bits (57), Expect = 7.7
Identities = 14/29 (48%), Positives = 15/29 (51%)
Frame = +3
Query: 94 VLLRDPTDENVVPVVEELQRRLPKVSSLH 122
VLLR P P LQRRLP+ LH
Sbjct: 336 VLLRRPHPRRHPPPNRRLQRRLPQAPHLH 422
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,533,514
Number of Sequences: 28460
Number of extensions: 66347
Number of successful extensions: 387
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 383
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 385
length of query: 393
length of database: 4,897,600
effective HSP length: 92
effective length of query: 301
effective length of database: 2,279,280
effective search space: 686063280
effective search space used: 686063280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0071.7


