

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0069.6
(167 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
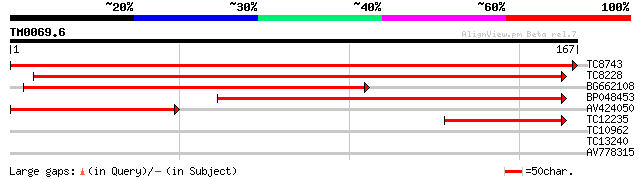
Score E
Sequences producing significant alignments: (bits) Value
TC8743 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxide ... 338 4e-94
TC8228 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid h... 255 2e-69
BG662108 135 3e-33
BP048453 133 2e-32
AV424050 100 1e-22
TC12235 similar to UP|GPX1_PEA (O24296) Phospholipid hydroperoxi... 59 6e-10
TC10962 similar to UP|Q9SV20 (Q9SV20) Beta-COP-like protein, par... 26 3.0
TC13240 26 4.0
AV778315 25 6.8
>TC8743 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxide
glutathione peroxidase , complete
Length = 680
Score = 338 bits (866), Expect = 4e-94
Identities = 167/167 (100%), Positives = 167/167 (100%)
Frame = +1
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS
Sbjct: 127 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 306
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK
Sbjct: 307 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 486
Query: 121 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQSS 167
GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQSS
Sbjct: 487 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQSS 627
>TC8228 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid
hydroperoxide glutathione peroxidase (PHGPx)
(Salt-associated protein) , partial (95%)
Length = 883
Score = 255 bits (652), Expect = 2e-69
Identities = 120/157 (76%), Positives = 137/157 (86%)
Frame = +2
Query: 8 SLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEILA 67
S+YDFTVKD RGNDV+L Y GKVL+IVNVASQCGLT +NY EL+ LYEKYKSKGLEIL
Sbjct: 191 SVYDFTVKDARGNDVNLGDYKGKVLLIVNVASQCGLTNSNYTELSQLYEKYKSKGLEILG 370
Query: 68 FPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGDG 127
FPCNQF QEPG N++IQ+ VCTRFK+EFPVFDKV+VNG +A PL+K+LK KGG+FGD
Sbjct: 371 FPCNQFGAQEPGDNEQIQEFVCTRFKAEFPVFDKVDVNGDSAAPLYKYLKSSKGGLFGDK 550
Query: 128 IKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
IKWNF+KFLV+KEG VVERYAPTTSP+ IEKDL KLL
Sbjct: 551 IKWNFSKFLVDKEGNVVERYAPTTSPLSIEKDLVKLL 661
>BG662108
Length = 410
Score = 135 bits (341), Expect = 3e-33
Identities = 60/102 (58%), Positives = 78/102 (75%)
Frame = +2
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
+ S+++F VKD RG DV+L+ Y GKVL++VNVAS+CG + NY +L LY +YK GLE
Sbjct: 104 SENSIHEFAVKDARGKDVNLNVYKGKVLLVVNVASKCGFAEANYTQLTQLYTRYKGSGLE 283
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNG 106
ILAFPCNQF +EPGT+ E QD VCTR+K+ +P+F KV VNG
Sbjct: 284 ILAFPCNQFLRKEPGTSQEAQDFVCTRYKAVYPIFGKVRVNG 409
>BP048453
Length = 527
Score = 133 bits (334), Expect = 2e-32
Identities = 58/103 (56%), Positives = 76/103 (73%)
Frame = -2
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKG 121
GLEILAFPCN F EPGT E + CTRFK+E+P+F KV V+G + PL++FLK +KG
Sbjct: 526 GLEILAFPCNPFLKPEPGTGQEAEPFACTRFKAEYPLFGKVRVHGADTAPLYRFLKPKKG 347
Query: 122 GIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
G G IKWNFT+FLV +EG V++RY TT+P+ +E D++K L
Sbjct: 346 GFLGSSIKWNFTQFLVEQEGNVLQRYGTTTTPLALENDIQKAL 218
>AV424050
Length = 274
Score = 100 bits (250), Expect = 1e-22
Identities = 50/50 (100%), Positives = 50/50 (100%)
Frame = +1
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKE 50
MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKE
Sbjct: 124 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKE 273
>TC12235 similar to UP|GPX1_PEA (O24296) Phospholipid hydroperoxide
glutathione peroxidase, chloroplast precursor (PHGPx) ,
partial (16%)
Length = 518
Score = 58.5 bits (140), Expect = 6e-10
Identities = 25/36 (69%), Positives = 30/36 (82%)
Frame = +2
Query: 129 KWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
KWNF KFL++K GKVVERY P TSP +IEKD+ +LL
Sbjct: 2 KWNFEKFLIDKNGKVVERYPPPTSPFQIEKDIHQLL 109
>TC10962 similar to UP|Q9SV20 (Q9SV20) Beta-COP-like protein, partial (36%)
Length = 1068
Score = 26.2 bits (56), Expect = 3.0
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = -1
Query: 123 IFGDGIKWNFTKFLVN 138
IFG+ IKW TK L+N
Sbjct: 273 IFGESIKWEVTKTLLN 226
>TC13240
Length = 518
Score = 25.8 bits (55), Expect = 4.0
Identities = 11/31 (35%), Positives = 17/31 (54%)
Frame = -1
Query: 130 WNFTKFLVNKEGKVVERYAPTTSPMKIEKDL 160
W FL+ K+ KVVE Y S M + +++
Sbjct: 155 WVLESFLIKKKTKVVEMYGNCKSMMDVFEEI 63
>AV778315
Length = 559
Score = 25.0 bits (53), Expect = 6.8
Identities = 8/19 (42%), Positives = 13/19 (68%)
Frame = -1
Query: 10 YDFTVKDIRGNDVSLSQYS 28
YDF D+ G D+++ QY+
Sbjct: 349 YDFLTPDLPGRDIAIPQYA 293
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,400,694
Number of Sequences: 28460
Number of extensions: 27018
Number of successful extensions: 78
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 78
length of query: 167
length of database: 4,897,600
effective HSP length: 84
effective length of query: 83
effective length of database: 2,506,960
effective search space: 208077680
effective search space used: 208077680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0069.6


