

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
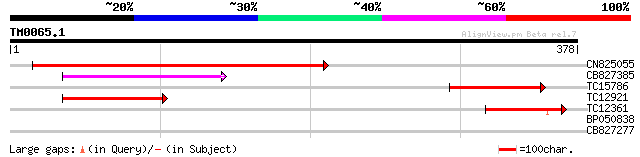
Query= TM0065.1
(378 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CN825055 401 e-112
CB827385 76 1e-14
TC15786 similar to UP|Q94II1 (Q94II1) ERD7 protein (Fragment), p... 67 6e-12
TC12921 64 5e-11
TC12361 similar to UP|Q94II1 (Q94II1) ERD7 protein (Fragment), p... 54 3e-08
BP050838 29 1.5
CB827277 26 9.5
>CN825055
Length = 593
Score = 401 bits (1030), Expect = e-112
Identities = 196/197 (99%), Positives = 196/197 (99%)
Frame = +2
Query: 16 MRTSTLVPIAEYSKPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSL 75
MRTSTLVPIAEYSKPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSL
Sbjct: 2 MRTSTLVPIAEYSKPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSL 181
Query: 76 ATIIKVGNGVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTFPEQCYGSMGLLE 135
ATIIKVGNGVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTFPEQCYGSMGLLE
Sbjct: 182 ATIIKVGNGVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTFPEQCYGSMGLLE 361
Query: 136 SFLKDHSCFSDLKTGKKGGLDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTN 195
SFLKDHSCFSDLKTGKKGGLDWE FAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTN
Sbjct: 362 SFLKDHSCFSDLKTGKKGGLDWEXFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTN 541
Query: 196 QVQSGGQTILTDNKNNG 212
QVQSGGQTILTDNKNNG
Sbjct: 542 QVQSGGQTILTDNKNNG 592
>CB827385
Length = 529
Score = 75.9 bits (185), Expect = 1e-14
Identities = 43/111 (38%), Positives = 66/111 (58%), Gaps = 2/111 (1%)
Frame = +2
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+ VL+ +PG ++L+++ +V LA G II +M+ + + + VG+ VQWPL KD
Sbjct: 173 EHVLVTVPGVILHLIEKDSSVHLASGDLNIISLMEGDRVVTVLASVGDQVQWPLAKDVSA 352
Query: 96 VKVDTLHYLFTLPV-KDGGS-EPLSYGVTFPEQCYGSMGLLESFLKDHSCF 144
VK+D HY FT+ V K+GG E L+YGVT + G L+ L+ + CF
Sbjct: 353 VKLDDSHYFFTIRVPKEGGEFEVLNYGVTVAAEGRGRGKELDEVLEKY-CF 502
>TC15786 similar to UP|Q94II1 (Q94II1) ERD7 protein (Fragment), partial
(17%)
Length = 600
Score = 66.6 bits (161), Expect = 6e-12
Identities = 33/64 (51%), Positives = 40/64 (61%)
Frame = +2
Query: 294 NKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIRK 353
+KV +A E A K +S +S T +V +RYGE+A EAT AGHA TAW KIRK
Sbjct: 5 SKVCDAVEVAGKNVMSTSSTVTTELVDHRYGEQAAEATSEGLNAAGHALGTAWAAFKIRK 184
Query: 354 AINP 357
AINP
Sbjct: 185 AINP 196
>TC12921
Length = 399
Score = 63.5 bits (153), Expect = 5e-11
Identities = 27/70 (38%), Positives = 45/70 (63%)
Frame = +3
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+E+L++IPG + L+D+ +VELA G + ++ + ++A +V + +QWPL KDE
Sbjct: 189 EEILLKIPGAILNLIDKDYSVELASGDLTVTRLRQGDNTIAVYARVADEIQWPLAKDETA 368
Query: 96 VKVDTLHYLF 105
VK+D HY F
Sbjct: 369 VKLDDSHYFF 398
>TC12361 similar to UP|Q94II1 (Q94II1) ERD7 protein (Fragment), partial
(15%)
Length = 412
Score = 54.3 bits (129), Expect = 3e-08
Identities = 27/57 (47%), Positives = 35/57 (61%), Gaps = 3/57 (5%)
Frame = +3
Query: 318 MVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIRKAINP---VTPASTAGGALRNS 371
+VS++YGEEA + T AGHA TAW V K+RKA+NP + P + A A R S
Sbjct: 57 LVSHKYGEEAAQVTNEGLDAAGHAIGTAWTVFKLRKALNPKSVIXPTTLAKAAARAS 227
>BP050838
Length = 474
Score = 28.9 bits (63), Expect = 1.5
Identities = 11/30 (36%), Positives = 20/30 (66%)
Frame = +2
Query: 16 MRTSTLVPIAEYSKPKNLIRQEVLIQIPGC 45
+R++TL+P+ P + + Q++LIQ P C
Sbjct: 380 LRSATLIPLQPPPTPLSSLHQQMLIQTPPC 469
>CB827277
Length = 546
Score = 26.2 bits (56), Expect = 9.5
Identities = 21/70 (30%), Positives = 31/70 (44%)
Frame = +2
Query: 113 GSEPLSYGVTFPEQCYGSMGLLESFLKDHSCFSDLKTGKKGGLDWEQFAPRVEDYNHFLA 172
G+ P S + +QC+ + ESF + F+ GG+ Q P + YNHF
Sbjct: 53 GTAPASRPMGNWQQCHSGV---ESFYGPPTGFTGPFIAPPGGIPGVQGPPHMVVYNHF-- 217
Query: 173 KAIAGGTGQI 182
A G GQ+
Sbjct: 218 -APVGQFGQV 244
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,076,512
Number of Sequences: 28460
Number of extensions: 56081
Number of successful extensions: 184
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 184
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 184
length of query: 378
length of database: 4,897,600
effective HSP length: 92
effective length of query: 286
effective length of database: 2,279,280
effective search space: 651874080
effective search space used: 651874080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0065.1


