

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0063.5
(435 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
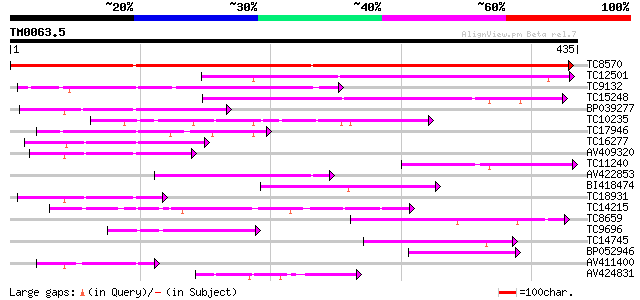
Score E
Sequences producing significant alignments: (bits) Value
TC8570 similar to UP|Q9FLM5 (Q9FLM5) N-hydroxycinnamoyl/benzoylt... 584 e-168
TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%) 171 2e-43
TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferas... 159 7e-40
TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A:... 155 9e-39
BP039277 109 1e-24
TC10235 100 5e-22
TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%) 94 6e-20
TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, par... 91 4e-19
AV409320 84 3e-17
TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transfera... 84 6e-17
AV422853 84 6e-17
BI418474 81 4e-16
TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl... 78 2e-15
TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%) 77 6e-15
TC8659 similar to UP|Q8VWP8 (Q8VWP8) Acyltransferase-like protei... 70 7e-13
TC9696 weakly similar to UP|Q8LAF8 (Q8LAF8) Fatty acid elongase-... 62 2e-10
TC14745 similar to UP|Q9LFB5 (Q9LFB5) Anthranilate N-benzoyltran... 60 7e-10
BP052946 60 9e-10
AV411400 59 1e-09
AV424831 55 2e-08
>TC8570 similar to UP|Q9FLM5 (Q9FLM5)
N-hydroxycinnamoyl/benzoyltransferase-like protein,
partial (91%)
Length = 1551
Score = 584 bits (1506), Expect = e-168
Identities = 285/432 (65%), Positives = 351/432 (80%)
Frame = +1
Query: 1 MANADMSNIGINVRQGEPTRVHPAEETEKGLYFLSNLDQNIAVPVRTVYCFKSSSRGNED 60
M N + + ++V+ EP V PAEET+KG+YFLSNLDQNIAV +RTVYCFK++ +GNE
Sbjct: 58 MENLNENGFQLSVKLSEPALVLPAEETKKGMYFLSNLDQNIAVIIRTVYCFKTAEKGNEK 237
Query: 61 AAHVIRVALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTK 120
A V++ AL K+LV YYP+AG+L IS EGKLIV+C T EGA+FVEAEA+C +E+IGD+TK
Sbjct: 238 AGEVVKSALKKVLVHYYPLAGRLSISPEGKLIVEC-TGEGALFVEAEANCSLEEIGDITK 414
Query: 121 PDPDSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAW 180
PDP +LGKL+Y+ PGA+ IL+MP + QVTKFKCGGF++GL + HCM DG+ AMEFVN+W
Sbjct: 415 PDPGTLGKLVYDIPGAKHILQMPPLVAQVTKFKCGGFSLGLCMNHCMFDGIGAMEFVNSW 594
Query: 181 SETARDLDLKTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCF 240
E AR L L PP LDR+ILKAR+PP IE H E+ +I D S+T LYE+E MLYRSFCF
Sbjct: 595 GEAARALPLSIPPILDRSILKARNPPKIEHLHQEFADIEDKSNTNTLYEDE-MLYRSFCF 771
Query: 241 DPKKLELLKKKATEDGVVKKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFV 300
DP+KL+ LK KA EDG ++ C+TFE LS FVW ART+ALK+ P+Q+TKLLFAVDGR F
Sbjct: 772 DPEKLKQLKMKAMEDGALESCTTFEVLSAFVWIARTKALKLLPEQETKLLFAVDGRKNFT 951
Query: 301 PPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRA 360
PP+P YFGN IVLT+SVC+AGE+ E LS +V L+ A+ +VTDSYMRS+IDYFEVTRA
Sbjct: 952 PPLPKGYFGNGIVLTNSVCQAGELSEKKLSHAVRLIQDAVKMVTDSYMRSAIDYFEVTRA 1131
Query: 361 RPSLTATLLITTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQDRKSINVLLGL 420
RPSL TLLITTW+RL FHTTDFGWGEP+ SGPV+LPEKEVILFLS+GQ+R++INVLLGL
Sbjct: 1132RPSLACTLLITTWSRLGFHTTDFGWGEPVLSGPVSLPEKEVILFLSHGQERRNINVLLGL 1311
Query: 421 PASAMETFEALM 432
PAS M+ F+ LM
Sbjct: 1312PASVMKIFQDLM 1347
>TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%)
Length = 1062
Score = 171 bits (434), Expect = 2e-43
Identities = 103/297 (34%), Positives = 168/297 (55%), Gaps = 11/297 (3%)
Frame = +3
Query: 148 QVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETAR--DLDLKTPPFLDRTILKARDP 205
QVT+F+CGGF++GL L HC+ DGL AM+F+ AW+ TA+ L + P DR + K R P
Sbjct: 48 QVTQFRCGGFSLGLRLCHCICDGLGAMQFLAAWAATAKSGSLVIDPKPCWDREMFKPRHP 227
Query: 206 PIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLKKKATEDGVVKKCSTFE 265
P+++F H E+ I + S+ + K + + + + LK A + C+TF+
Sbjct: 228 PMVKFPHMEFMRIEEGSNLTITLWQTKPVQKCYRIQREFQNYLKTLA-QPSDAAGCTTFD 404
Query: 266 ALSGFVWRARTEALKMEP-DQKTKLLFAVDGRTRF-VPPIPDRYFGNAIVLTHSVCKAGE 323
A++ +WR+ +AL + P D +L F+V+ R + PP+ + ++GN + + + E
Sbjct: 405 AMAAHIWRSWVKALDVRPLDYTLRLTFSVNARPKLRNPPLREGFYGNVVCVACTTSSVSE 584
Query: 324 IVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARP-SLTATLLITTWTRLS-FHTT 381
+V L + LV A V++ Y+RS++DY +V R + L IT WTR S + +
Sbjct: 585 LVHGQLPETTRLVREARQSVSEEYLRSTVDYVDVDRPKQLEFGGKLTITQWTRFSMYKSA 764
Query: 382 DFGWGEPLCSGPVTL-PEKEVILFLSNGQDRK---SINVLLGLPASAMETF-EALMM 433
DFGWG+PL +GP+ L P +V +FL G+ S+ V + LP SA + F +AL++
Sbjct: 765 DFGWGKPLYAGPIDLTPTPQVCVFLPEGEAADCSGSMIVCICLPESAAQKFTQALLL 935
>TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(53%)
Length = 878
Score = 159 bits (403), Expect = 7e-40
Identities = 97/253 (38%), Positives = 144/253 (56%), Gaps = 3/253 (1%)
Frame = +1
Query: 7 SNIGINVRQGEPTRVHPAEETEKGLYFLSNLDQNIAVP---VRTVYCFKSSSRGNEDAAH 63
S + I VR E T V PAEE + + SN+D + VP +VY ++++ N A
Sbjct: 142 SEMIITVR--ESTMVRPAEEVPQRTVWNSNVD--LVVPNFHTPSVYFYRANGASNFFDAK 309
Query: 64 VIRVALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDP 123
V++ ALSK+LVP+YPMAG+L +G++ +DC+ +G +FVEA+ I+D GD
Sbjct: 310 VLKEALSKVLVPFYPMAGRLRRDDDGRVEIDCDG-QGVLFVEADTGAVIDDFGDFAPTLQ 486
Query: 124 DSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSET 183
+LI +R I PL+ +QVT FKCGG ++G+ + H + DG S + F+N WS+
Sbjct: 487 QR--QLIPGVDYSRGIESYPLLVLQVTHFKCGGVSLGVGMQHHVADGASGLHFINTWSDV 660
Query: 184 ARDLDLKTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDPK 243
AR LD+ PPF+DRT+L+ARDPP F+H EY ++ E + F
Sbjct: 661 ARGLDVSIPPFIDRTLLRARDPPRPAFEHIEYKPPPSMAKPGSDSSEVSI----FKLTRD 828
Query: 244 KLELLKKKATEDG 256
+L LK K+ E+G
Sbjct: 829 QLNTLKAKSREEG 867
>TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A: benzyl
alcohol benzoyl transferase, partial (52%)
Length = 1085
Score = 155 bits (393), Expect = 9e-39
Identities = 91/288 (31%), Positives = 144/288 (49%), Gaps = 8/288 (2%)
Frame = +1
Query: 149 VTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDLDLKTP-PFLDRTILKARDPPI 207
VT+ KCGGF L H M DG + F+ A E AR + + P R +L ARDPP
Sbjct: 13 VTRLKCGGFIFALRFNHVMTDGAGIVHFMYAVVEIARGANEPSILPVWQRELLHARDPPR 192
Query: 208 IEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLKKKATEDGVVKKCSTFEAL 267
+ H EY+++ D + T + + +SF F P ++ +++ + + +T+E L
Sbjct: 193 VTHNHREYEQLTDDTITDPILTGTDFVQQSFFFGPAEIAAIRRHLPHH-LDTESTTYEVL 369
Query: 268 SGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIVEN 327
+ ++WR RT+AL+++P Q+ +++ D R +F PP P Y+GN +V AGE+ E
Sbjct: 370 TSYIWRCRTKALQLDPSQEVRMMCITDARGKFNPPFPTGYYGNCFAFPAAVATAGELCEK 549
Query: 328 PLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARPSLTA--TLLITTWTRLSFHTTDFGW 385
PL +V L+ A +++ YM S D VT +P T + ++ T F DFGW
Sbjct: 550 PLEHAVRLIKKASGEMSEEYMHSLADLM-VTEGKPLFTVVRSCVVLDTTYAGFRELDFGW 726
Query: 386 GEPLC-----SGPVTLPEKEVILFLSNGQDRKSINVLLGLPASAMETF 428
G+ + +G P + N Q + I VL+ LPA M F
Sbjct: 727 GKAVYGGLAQAGAGAFPAVNFHVPSQNAQGEEGILVLVCLPAKIMSVF 870
>BP039277
Length = 522
Score = 109 bits (272), Expect = 1e-24
Identities = 62/167 (37%), Positives = 92/167 (54%), Gaps = 4/167 (2%)
Frame = +1
Query: 8 NIGINVRQGEPTRVHPAEETEKGLYFLSNLDQN----IAVPVRTVYCFKSSSRGNEDAAH 63
++ V++ +P V PA T + LS++D +P +Y + S ++D
Sbjct: 16 SLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSM-ADKDPVQ 192
Query: 64 VIRVALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDP 123
IR ALS+ LV YYP AG+L KL+VDC T EG +F+EA AD + + G+ P
Sbjct: 193 AIRQALSRTLVYYYPFAGRLKEGLGRKLMVDC-TGEGVMFIEANADVSLVEFGETPLPPF 369
Query: 124 DSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDG 170
+L+Y+ PG+ +L+ PL+ +QVT+ KCGGF M L H M DG
Sbjct: 370 PCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDG 510
>TC10235
Length = 1116
Score = 100 bits (249), Expect = 5e-22
Identities = 86/282 (30%), Positives = 137/282 (48%), Gaps = 19/282 (6%)
Frame = +1
Query: 63 HVIRVALSKILVPYYPMAGKLVIS--------TEGKLIVDCNTMEGAVFVEAEADCDIED 114
H ++ LS IL + P G+L+I+ T + CN GA+FV A A+
Sbjct: 247 HHLKHTLSTILTFFPPFTGRLIITNHHDNNNNTTVTCHITCNNA-GALFVHAAAEKT--S 417
Query: 115 IGDLTKPD-PDSLGKLIYNTPGARSI--LEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGL 171
I D+ +P S+ + G ++ + PL+ VQVT+ G F +G + H + DG
Sbjct: 418 IADIIRPGYVPSIVHSFFPLNGVKNYQGMSQPLLAVQVTELLDGVF-IGFTMNHSVADGK 594
Query: 172 SAMEFVNAWSETAR---DLDLKTPPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLY 228
+ +F+N+W E +R D +KTPP L+R DPPI F +E D
Sbjct: 595 AFWQFINSWGELSRGNSDKLMKTPP-LERWFPNGIDPPI-HFPSFTKEEEKKKQDNSDPV 768
Query: 229 EEEKMLYRSFCFDPKKLELLKKKAT---EDGVVKK--CSTFEALSGFVWRARTEALKMEP 283
+ L F F +KL LK KA ++G+ + S+ +AL +WR+ A ++ P
Sbjct: 769 PQLPPLC-IFHFSKEKLAELKAKANAEVDNGMHENRVISSLQALLTHIWRSVIRAQRVNP 945
Query: 284 DQKTKLLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIV 325
+++T + +D R R PP+P+ Y GNA + KAGE++
Sbjct: 946 EKETDYMLIIDARQRMQPPLPENYLGNAGMAGIVKLKAGELL 1071
>TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%)
Length = 699
Score = 93.6 bits (231), Expect = 6e-20
Identities = 61/187 (32%), Positives = 103/187 (54%), Gaps = 6/187 (3%)
Frame = +3
Query: 21 VHPAEETEKGLYFLSNLDQNIAVPVRTVYCFKSSSRGNEDAAHVIRVALSKILVPYYPMA 80
V P + T +LS++DQ ++ T + ++ ++ ++ +LSKILV YYP+A
Sbjct: 144 VPPNQPTPH*HLWLSDMDQ-VSRQRHTPIVYIYKAKHDQYCIERMKDSLSKILVHYYPVA 320
Query: 81 GKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPD--PDSLGKLIYNTPGARS 138
G+ + G++ ++CN +G + +EAE + + D GD + D + + + YN P
Sbjct: 321 GRFSFTENGRMEINCNA-KGVILLEAETEKTMADFGDFSPSDSTKELVPTIDYNHP---- 485
Query: 139 ILEMPLMTVQVTKFKC--GGFTMGLNLIHCMKDGLSAMEFVNAWSETAR--DLDLKTPPF 194
I E+PL+ VQ+T+F GG G+ +H + DGL F N+W++ AR L+ P
Sbjct: 486 IEELPLLAVQLTRFNGGEGGLADGVAWVHTLSDGLGCTRFFNSWAKIARGDTLEPHEKPS 665
Query: 195 LDRTILK 201
LDRT+LK
Sbjct: 666 LDRTLLK 686
>TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, partial (32%)
Length = 547
Score = 90.9 bits (224), Expect = 4e-19
Identities = 55/147 (37%), Positives = 82/147 (55%), Gaps = 5/147 (3%)
Frame = +1
Query: 12 NVRQGEPTRVHPAEETEKGLYFLSNLDQNIA----VPVRTVYCFKSSSRGNEDAAHVIRV 67
+VR+ EP V PA T LS++D VP+ Y + S G +D +IR
Sbjct: 112 SVRRREPELVAPAVSTPHETKHLSDIDDQAGLRANVPIIQFYRNEPSMAG-KDPVDIIRN 288
Query: 68 ALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLG 127
A++K LV YYP+AG+L + G L+VDCN EG +F+EA+AD E GD KP
Sbjct: 289 AVAKALVFYYPLAGRLREAAGGNLVVDCNE-EGVMFIEADADITFEQFGDTLKPPFPCFQ 465
Query: 128 KLIYNTPGARS-ILEMPLMTVQVTKFK 153
+L++ PG+ ++ P++ +QVT+ K
Sbjct: 466 ELLHEAPGSEGVVINSPMILIQVTRLK 546
>AV409320
Length = 392
Score = 84.3 bits (207), Expect = 3e-17
Identities = 51/132 (38%), Positives = 72/132 (53%), Gaps = 4/132 (3%)
Frame = +3
Query: 16 GEPTRVHPAEETEKGLYFLSNLDQN----IAVPVRTVYCFKSSSRGNEDAAHVIRVALSK 71
GEP V PAE T + + LS++D +PV Y + S G +D IR AL+K
Sbjct: 3 GEPELVGPAEATPREVKLLSDIDDQDGLRFQIPVAQFYRYNPSMAG-KDPVDAIRKALAK 179
Query: 72 ILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKLIY 131
LV YYP AG+L KL+VDC T EG +F+EA+AD + + GD + + +L+Y
Sbjct: 180 TLVFYYPFAGRLREGPGRKLMVDC-TAEGVLFIEADADVTLNEFGDNLQTPFPCMEELLY 356
Query: 132 NTPGARSILEMP 143
PG+ +L P
Sbjct: 357 EVPGSDEMLNTP 392
>TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(31%)
Length = 528
Score = 83.6 bits (205), Expect = 6e-17
Identities = 42/146 (28%), Positives = 74/146 (49%), Gaps = 11/146 (7%)
Frame = +1
Query: 301 PPIPDRYFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRA 360
PP+P YFGN I + AG+++ P+ ++ +H A+ + + Y+RS++D+ E+
Sbjct: 28 PPLPQGYFGNVIFTATPIAMAGDLMSKPIWYAASRIHDALLRMDNEYLRSALDFLEL--- 198
Query: 361 RPSLTA-----------TLLITTWTRLSFHTTDFGWGEPLCSGPVTLPEKEVILFLSNGQ 409
+P L A L IT+W RL H DFGWG P+ GP + + + + +
Sbjct: 199 QPDLRALVRGAHTFRCPNLGITSWVRLPIHDADFGWGRPIFMGPGGIAYEGLSFIIPSPV 378
Query: 410 DRKSINVLLGLPASAMETFEALMMQV 435
+ S++V + L M+ F+ L +
Sbjct: 379 NDGSLSVAIALQPEQMKVFQELFYDI 456
>AV422853
Length = 456
Score = 83.6 bits (205), Expect = 6e-17
Identities = 46/139 (33%), Positives = 74/139 (53%), Gaps = 1/139 (0%)
Frame = +1
Query: 112 IEDIGDLTKPDPDSLGKLIYNTPGARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGL 171
+ GD + + +L+Y PG+ +L PL+ +QVT+ KCGGF + L H M D +
Sbjct: 7 LNQFGDNLQTPFPCMDELLYEVPGSEEMLNTPLLLIQVTRLKCGGFIFAIRLNHTMCDAV 186
Query: 172 SAMEFVNAWSETARDLDLKT-PPFLDRTILKARDPPIIEFKHHEYDEIVDLSDTKKLYEE 230
++F++A E AR + + P R IL ARD P + H EYDE V + ++
Sbjct: 187 GLVQFLSAIGEMARGMPQPSILPVWCREILSARDSPRVTCTHPEYDEQVPYPKETTIPQD 366
Query: 231 EKMLYRSFCFDPKKLELLK 249
+ M++ SF F P +L ++
Sbjct: 367 D-MVHESFFFGPNELATIR 420
>BI418474
Length = 479
Score = 80.9 bits (198), Expect = 4e-16
Identities = 49/143 (34%), Positives = 77/143 (53%), Gaps = 5/143 (3%)
Frame = +3
Query: 193 PFLDRTILKARDPPII-EFKHHEYDEI-VDLSDTKKLYEEEKMLYRSFCF-DPKKLELLK 249
PFLDRT+LK PP F+H E + ++L + E K + P ++E LK
Sbjct: 42 PFLDRTLLKFSQPPSAPRFEHKELKPLPLNLGSSDCSVERNKKVTSELLILTPLQVEKLK 221
Query: 250 KKATEDGVV--KKCSTFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRY 307
K+A + + + S +E + +WR ++A +++ +Q T + F V+ R+R VPP+P Y
Sbjct: 222 KEANGNVLEGSRPYSRYEVIGAHIWRCASKARELDQNQPTVVRFNVENRSRMVPPLPPSY 401
Query: 308 FGNAIVLTHSVCKAGEIVENPLS 330
FGNA+ T + GEI PLS
Sbjct: 402 FGNALTQTAATGYVGEITSKPLS 470
>TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl alcohol
benzoyl transferase, partial (26%)
Length = 446
Score = 78.2 bits (191), Expect = 2e-15
Identities = 45/119 (37%), Positives = 71/119 (58%), Gaps = 4/119 (3%)
Frame = +3
Query: 7 SNIGINVRQGEPTRVHPAEETEKGLYFLSNLDQN----IAVPVRTVYCFKSSSRGNEDAA 62
+++ VR+ EP V P++ T + L LS++D +PV Y + S G +D
Sbjct: 78 ASLAFTVRRCEPELVAPSQPTPRELKLLSDIDDQEGLRFQIPVIQFYRYDPSMTG-KDPV 254
Query: 63 HVIRVALSKILVPYYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKP 121
VIR AL+K LV YYP+AG+L+ + KL+V+C T +G +F+EA+AD ++ GD +P
Sbjct: 255 EVIRKALAKTLVFYYPLAGRLMEGPDRKLMVNC-TADGVLFIEADADVTMKQFGDALQP 428
>TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%)
Length = 1096
Score = 77.0 bits (188), Expect = 6e-15
Identities = 74/285 (25%), Positives = 125/285 (42%), Gaps = 5/285 (1%)
Frame = +1
Query: 31 LYFLSNLDQNIAVPVRTVYCFKSSSRGNEDAAHVIRVALSKILVPYYPMAGKLVISTEGK 90
L F+ N+ +PV Y S + + ++ +LS++L YYP AG+L +
Sbjct: 169 LSFIDNMMYRNYIPVALFYHPNESDK--QSIISNLKNSLSEVLTRYYPFAGRL----RDQ 330
Query: 91 LIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKLIY--NTPGARSILEMPLMTVQ 148
L ++CN EG F E + I L P+ ++L +L++ P + + + VQ
Sbjct: 331 LSIECND-EGVPFRVTEIKGEQSTI--LQNPN-ETLLRLLFPDKLPWKVTEYDESIAAVQ 498
Query: 149 VTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDLDLKTPPFLDRTILKARDPPII 208
+ F CGG + + H M D + + FVN W+ ++ L + L + +
Sbjct: 499 INFFSCGGLAITACVCHKMGDATTTLNFVNDWAAMTKE----KGEALTQLSLPLLNGGVS 666
Query: 209 EFKHHE---YDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLKKKATEDGVVKKCSTFE 265
F H + + EIV K + R F F P K+ LK+ T G V+ + E
Sbjct: 667 VFPHKDMPCFPEIVFGKGNK-----SNAVCRRFVFPPSKINSLKEMVTSHG-VQNPTRVE 828
Query: 266 ALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGN 310
++ +++ AL + D KT V R R PP+P++ GN
Sbjct: 829 VITAWIYMRAVHALGLTFD-KTSFRQVVSLRRRMTPPLPNKSVGN 960
>TC8659 similar to UP|Q8VWP8 (Q8VWP8) Acyltransferase-like protein
(Fragment), partial (42%)
Length = 866
Score = 70.1 bits (170), Expect = 7e-13
Identities = 51/174 (29%), Positives = 80/174 (45%), Gaps = 6/174 (3%)
Frame = +3
Query: 262 STFEALSGFVWRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKA 321
STF+ALS +WR T A ++P+ T VD R R PP+P+ YFGN I +V
Sbjct: 3 STFQALSTHIWRHVTHARSLKPEDYTVFTVFVDCRKRVDPPMPEAYFGNLIQAIFTVTAV 182
Query: 322 GEIVENPLSFSVGLVHSAIDI----VTDSYMRSSIDYFEVTRARPSLTATLLITTWTRLS 377
G + +P F ++ AI+ D + ++ + + + + + + R
Sbjct: 183 GLLSAHPPQFGATMIQKAIEAHDAKAIDERNKDWESAPKIFQFKDAGINCVTVGSSPRFK 362
Query: 378 FHTTDFGWGEP--LCSGPVTLPEKEVILFLSNGQDRKSINVLLGLPASAMETFE 429
+ DFGWG+P + SGP + + L+ R SI+V L L AM E
Sbjct: 363 VYDIDFGWGKPEIVRSGPNNKFDGMIYLYPGKSGGR-SIDVELTLEPEAMGRLE 521
>TC9696 weakly similar to UP|Q8LAF8 (Q8LAF8) Fatty acid elongase-like
protein (cer2-like), partial (23%)
Length = 881
Score = 62.0 bits (149), Expect = 2e-10
Identities = 35/117 (29%), Positives = 60/117 (50%)
Frame = +3
Query: 76 YYPMAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDIGDLTKPDPDSLGKLIYNTPG 135
YY G+ S G+ + CN G+ F+EA+ +++ L D L+
Sbjct: 306 YYFTCGRFRRSESGRPFIKCNDC-GSRFIEAKCKKTLDEW--LAMKDWPLYKLLVSQQVI 476
Query: 136 ARSILEMPLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSETARDLDLKTP 192
+ P + Q+T+FKCGG ++GL+ H + D LSA +F+N+W + ++ +KTP
Sbjct: 477 GPELSFSPSVLFQLTQFKCGGISLGLSWAHVLGDPLSASDFINSWGQVMSNMGMKTP 647
>TC14745 similar to UP|Q9LFB5 (Q9LFB5) Anthranilate
N-benzoyltransferase-like protein (AT5g01210/F7J8_190),
partial (37%)
Length = 842
Score = 60.1 bits (144), Expect = 7e-10
Identities = 38/122 (31%), Positives = 58/122 (47%), Gaps = 4/122 (3%)
Frame = +1
Query: 272 WRARTEALKMEPDQKTKLLFAVDGRTRFVPPIPDRYFGNAIVLTHSVCKAGEIVENPLSF 331
WR+ T A K++ + + AV+ R R P + YFGNAI +V AG+I+ L F
Sbjct: 1 WRSVTRARKLDAAKTSTFRMAVNCRHRLEPKMDPFYFGNAIQSIPTVASAGDILSRDLRF 180
Query: 332 SVGLVHSAIDIVTDSYMRSSIDYFEVTRARPSL----TATLLITTWTRLSFHTTDFGWGE 387
L+H + D+ +R I+ +E L A + + + R + DFGWG
Sbjct: 181 CADLLHQNVVAHDDATVRCGIEGWEKAPRLFPLGNFDGAMITMGSSPRFPMYDNDFGWGR 360
Query: 388 PL 389
PL
Sbjct: 361 PL 366
>BP052946
Length = 559
Score = 59.7 bits (143), Expect = 9e-10
Identities = 30/87 (34%), Positives = 42/87 (47%), Gaps = 1/87 (1%)
Frame = -1
Query: 307 YFGNAIVLTHSVCKAGEIVENPLSFSVGLVHSAIDIVTDSYMRSSIDYFEVTRARPSLTA 366
Y+GN V G++ N ++V LV A T+ YM S D+ R T
Sbjct: 553 YYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMXDFLVANRRCLFTTV 374
Query: 367 -TLLITTWTRLSFHTTDFGWGEPLCSG 392
+ +++ TR H TDFGWGEP+C G
Sbjct: 373 RSCIVSDLTRFKLHETDFGWGEPVCGG 293
>AV411400
Length = 417
Score = 59.3 bits (142), Expect = 1e-09
Identities = 39/97 (40%), Positives = 55/97 (56%), Gaps = 2/97 (2%)
Frame = +2
Query: 21 VHPAEETEKGLYFLSNLDQN--IAVPVRTVYCFKSSSRGNEDAAHVIRVALSKILVPYYP 78
V PA+ET LS +D+ + RT++ F R +A VIR ALS LVPYYP
Sbjct: 59 VKPAKETPLTTLDLSVMDRLPVLRCNARTLHVF----RHGPEATRVIREALSLALVPYYP 226
Query: 79 MAGKLVISTEGKLIVDCNTMEGAVFVEAEADCDIEDI 115
+AG+L+ S L ++C+ EG +VEA ADC + +
Sbjct: 227 LAGRLIESKPRCLQIECSG-EGVWYVEASADCTLHSV 334
>AV424831
Length = 415
Score = 55.5 bits (132), Expect = 2e-08
Identities = 44/141 (31%), Positives = 64/141 (45%), Gaps = 13/141 (9%)
Frame = +3
Query: 143 PLMTVQVTKFKCGGFTMGLNLIHCMKDGLSAMEFVNAWSE-------TARDLDLKTPPFL 195
PL+T++VT+ G F +G ++ HC+ DG + F N WSE T D + P
Sbjct: 21 PLLTIKVTELLDGVF-IGCSMNHCVGDGTAYWNFFNTWSEIFQAKAQTGHD-EFHYVPIS 194
Query: 196 DRTILKARDPP------IIEFKHHEYDEIVDLSDTKKLYEEEKMLYRSFCFDPKKLELLK 249
I PP + F+HH DE + YE K+ R F F + + LK
Sbjct: 195 HHPIHNR*FPPGCNRLINLPFRHH--DEFIG------RYESTKLRERIFHFSAESIAKLK 350
Query: 250 KKATEDGVVKKCSTFEALSGF 270
KA + K S+F++LS F
Sbjct: 351 AKANSESKSNKISSFQSLSAF 413
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,100,677
Number of Sequences: 28460
Number of extensions: 91030
Number of successful extensions: 483
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 456
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 457
length of query: 435
length of database: 4,897,600
effective HSP length: 93
effective length of query: 342
effective length of database: 2,250,820
effective search space: 769780440
effective search space used: 769780440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0063.5


