

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0062.3
(108 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
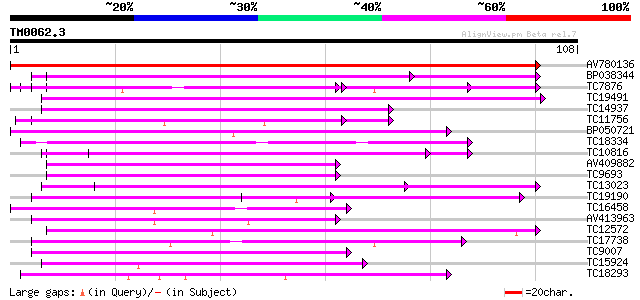
Score E
Sequences producing significant alignments: (bits) Value
AV780136 195 1e-51
BP038344 54 4e-09
TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-bi... 54 5e-09
TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Ar... 49 1e-07
TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5,... 46 8e-07
TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snR... 46 1e-06
BP050721 45 2e-06
TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1 (AT3... 44 4e-06
TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subun... 43 9e-06
AV409882 42 2e-05
TC9693 similar to UP|COPP_MOUSE (O55029) Coatomer beta' subunit ... 42 2e-05
TC13023 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragm... 41 3e-05
TC19190 weakly similar to PIR|T46032|T46032 WD-40 repeat regulat... 41 3e-05
TC16458 similar to UP|O48679 (O48679) F3I6.5 protein, partial (20%) 41 3e-05
AV413963 41 3e-05
TC12572 similar to PIR|T46032|T46032 WD-40 repeat regulatory pro... 39 1e-04
TC17738 similar to UP|O48679 (O48679) F3I6.5 protein, partial (8%) 39 2e-04
TC9007 similar to UP|Q9SSS7 (Q9SSS7) F6D8.2 protein (At1g52730),... 39 2e-04
TC15924 weakly similar to GB|BAC21577.1|24060129|AP005438 G-prot... 37 4e-04
TC18293 similar to UP|PWP1_HUMAN (Q13610) Periodic tryptophan pr... 37 7e-04
>AV780136
Length = 558
Score = 195 bits (495), Expect = 1e-51
Identities = 90/101 (89%), Positives = 98/101 (96%)
Frame = -1
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
MNEGGT+LVSGGTEKV+RVWDPRSGSKT+KL+GH DNIRALLLDSTGR+CLSGSSDSMIR
Sbjct: 549 MNEGGTVLVSGGTEKVIRVWDPRSGSKTLKLRGHADNIRALLLDSTGRYCLSGSSDSMIR 370
Query: 61 LWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
LWD+GQ RCLH+YAVHTDSVWALAST TFSHVYSGGRDFS+
Sbjct: 369 LWDIGQPRCLHTYAVHTDSVWALASTPTFSHVYSGGRDFSL 247
>BP038344
Length = 525
Score = 53.9 bits (128), Expect = 4e-09
Identities = 28/97 (28%), Positives = 47/97 (47%)
Frame = +2
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
GTLL S +K + +W + S +L GH++ I L S + S S D +R+WD
Sbjct: 134 GTLLASASLDKTLIIWSSATLSLLHRLTGHSEGISDLAWSSDSHYICSASDDRTLRIWDA 313
Query: 65 GQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
C+ + HT +V+ + +++ SG D +V
Sbjct: 314 TGGDCVKTLRGHTHAVFCVNFNPQSNYIVSGSFDETV 424
Score = 53.1 bits (126), Expect = 7e-09
Identities = 20/70 (28%), Positives = 37/70 (52%)
Frame = +2
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+ S ++ +R+WD G L+GHT + + + + +SGS D +R+W++
Sbjct: 269 ICSASDDRTLRIWDATGGDCVKTLRGHTHAVFCVNFNPQSNYIVSGSFDETVRVWEVKTG 448
Query: 68 RCLHSYAVHT 77
RC+H+ HT
Sbjct: 449 RCIHTIIAHT 478
Score = 26.6 bits (57), Expect = 0.67
Identities = 12/46 (26%), Positives = 23/46 (49%)
Frame = +2
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTG 47
N +VSG ++ VRVW+ ++G + HT + ++ + G
Sbjct: 377 NPQSNYIVSGSFDETVRVWEVKTGRCIHTIIAHTMPVTSVHFNRDG 514
>TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1379
Score = 53.5 bits (127), Expect = 5e-09
Identities = 29/104 (27%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Frame = +3
Query: 3 EGGTLLVSGGTEKVVRVW-----DPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDS 57
+ ++V+ +K + +W D G +L GH+ ++ ++L S G+F LSGS D
Sbjct: 147 DNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDG 326
Query: 58 MIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSV 101
+RLWDL + HT V ++A + + S RD ++
Sbjct: 327 ELRLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTI 458
Score = 45.8 bits (107), Expect = 1e-06
Identities = 23/90 (25%), Positives = 40/90 (43%), Gaps = 9/90 (10%)
Frame = +3
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+VS ++ V+VW+ + L GH+ + + + G C SG D +I LWDL +
Sbjct: 567 IVSASWDRTVKVWNLTNCKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEG 746
Query: 68 R---------CLHSYAVHTDSVWALASTST 88
+ +H+ + W A+T T
Sbjct: 747 KRLYSLDAGSIIHALCFSPNRYWLCAATET 836
Score = 40.4 bits (93), Expect = 5e-05
Identities = 17/63 (26%), Positives = 33/63 (51%)
Frame = +3
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
++ G +SG + +R+WD +G+ + GHT ++ ++ R +S S D I+
Sbjct: 282 LSSDGQFALSGSWDGELRLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTIK 461
Query: 61 LWD 63
LW+
Sbjct: 462 LWN 470
Score = 36.6 bits (83), Expect = 7e-04
Identities = 19/60 (31%), Positives = 32/60 (52%)
Frame = +3
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G+L SGG + V+ +WD G + L +I L S R+ L ++++ I++WDL
Sbjct: 684 GSLCASGGKDGVILLWDLAEGKRLYSL--DAGSIIHALCFSPNRYWLCAATETSIKIWDL 857
>TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Arabidopsis
thaliana;}, partial (39%)
Length = 527
Score = 48.9 bits (115), Expect = 1e-07
Identities = 28/96 (29%), Positives = 44/96 (45%)
Frame = +1
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
LL + + V ++D + + GH + + + G +GSSD +RLWDL
Sbjct: 61 LLFTASDDGNVHMYDAEGKALVGTMSGHASWVLCVDVSPDGGAIATGSSDRTVRLWDLAM 240
Query: 67 QRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSVS 102
+ + + + HTD VW +A GGR SVS
Sbjct: 241 RASVQTMSNHTDQVWGVAFRPPGGTDVRGGRLASVS 348
Score = 30.8 bits (68), Expect = 0.036
Identities = 17/65 (26%), Positives = 28/65 (42%), Gaps = 6/65 (9%)
Frame = +1
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLL------DSTGRFCLSGSSDSM 58
G + +G +++ VR+WD + + HTD + + D G S S D
Sbjct: 181 GGAIATGSSDRTVRLWDLAMRASVQTMSNHTDQVWGVAFRPPGGTDVRGGRLASVSDDKS 360
Query: 59 IRLWD 63
I L+D
Sbjct: 361 ISLYD 375
>TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5, TAC
clone:K8K14 (AT5g67320/K8K14_4), partial (19%)
Length = 1089
Score = 46.2 bits (108), Expect = 8e-07
Identities = 19/67 (28%), Positives = 32/67 (47%)
Frame = +1
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
+L S + V++WD G L GH D + ++ G + SGS D I +W L +
Sbjct: 52 VLASASFDSTVKLWDVEVGKLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWSLKE 231
Query: 67 QRCLHSY 73
+ + +Y
Sbjct: 232 GKIVKTY 252
Score = 35.8 bits (81), Expect = 0.001
Identities = 18/57 (31%), Positives = 32/57 (55%)
Frame = +1
Query: 52 SGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRDFSVSAVFLKK 108
S S DS ++LWD+ + ++S H D V+++A + ++ SG D S+ LK+
Sbjct: 61 SASFDSTVKLWDVEVGKLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWSLKE 231
>TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snRNP-specific
40 kDa protein (hPrp8-binding) {Homo sapiens;} , partial
(43%)
Length = 719
Score = 45.8 bits (107), Expect = 1e-06
Identities = 22/70 (31%), Positives = 36/70 (51%), Gaps = 1/70 (1%)
Frame = +1
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTG-RFCLSGSSDSMIRLWD 63
GT +VS +K VRVWD +G + K+ H + + G +SGS D +LWD
Sbjct: 469 GTQIVSASPDKTVRVWDVETGKQVKKMVEHLSYVNSCCPSRRGPPLVVSGSDDGTAKLWD 648
Query: 64 LGQQRCLHSY 73
+ Q+ + ++
Sbjct: 649 MRQRGSIQTF 678
Score = 40.0 bits (92), Expect = 6e-05
Identities = 20/64 (31%), Positives = 34/64 (52%), Gaps = 1/64 (1%)
Frame = +1
Query: 2 NEGGTLLVSGGTEKVVRVWDPRSGSKT-MKLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
N GT++ SG ++ + +W+ K M LKGH + + L + G +S S D +R
Sbjct: 331 NPAGTVIASGSHDREIFLWNVHGECKNFMVLKGHKNAVLDLHWTTDGTQIVSASPDKTVR 510
Query: 61 LWDL 64
+WD+
Sbjct: 511 VWDV 522
Score = 28.5 bits (62), Expect = 0.18
Identities = 22/77 (28%), Positives = 34/77 (43%), Gaps = 1/77 (1%)
Frame = +1
Query: 26 SKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL-GQQRCLHSYAVHTDSVWALA 84
S M L GH I + + G SGS D I LW++ G+ + H ++V L
Sbjct: 277 SPIMLLTGHQSVIYTMKFNPAGTVIASGSHDREIFLWNVHGECKNFMVLKGHKNAVLDLH 456
Query: 85 STSTFSHVYSGGRDFSV 101
T+ + + S D +V
Sbjct: 457 WTTDGTQIVSASPDKTV 507
>BP050721
Length = 562
Score = 44.7 bits (104), Expect = 2e-06
Identities = 26/86 (30%), Positives = 40/86 (46%), Gaps = 2/86 (2%)
Frame = -1
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRAL--LLDSTGRFCLSGSSDSM 58
M+ G +++G + VR+ D SG + KGHT+ L L +T GS D
Sbjct: 487 MSNDGNCILAGCLDSTVRLLDRTSGELLQEYKGHTNKSYKLDCCLTNTDAHVAGGSEDGF 308
Query: 59 IRLWDLGQQRCLHSYAVHTDSVWALA 84
I WDL + S+ HT V +++
Sbjct: 307 IYFWDLVDAYVVSSFRAHTSVVTSVS 230
Score = 28.1 bits (61), Expect = 0.23
Identities = 17/63 (26%), Positives = 26/63 (40%), Gaps = 1/63 (1%)
Frame = -1
Query: 9 VSGGTEK-VVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
V+GG+E + WD + HT + ++ LS S D IR+W QQ
Sbjct: 334 VAGGSEDGFIYFWDLVDAYVVSSFRAHTSVVTSVSYHPKENCMLSASVDGTIRVWKT*QQ 155
Query: 68 RCL 70
+
Sbjct: 154 NAV 146
>TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1
(AT3g18860/MCB22_3), partial (21%)
Length = 595
Score = 43.9 bits (102), Expect = 4e-06
Identities = 28/86 (32%), Positives = 41/86 (47%)
Frame = +2
Query: 3 EGGTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLW 62
+GG + SGG + +V VWD +G K LKGH + ++ D +S S D +R W
Sbjct: 329 QGG--VASGGMDTLVLVWDLSTGEKAHTLKGHQLQVTSIAFDDGD--VVSASVDCTLRRW 496
Query: 63 DLGQQRCLHSYAVHTDSVWALASTST 88
GQ C ++ H V A+ T
Sbjct: 497 KNGQ--CTETWEAHKAPVQAVIKLPT 568
>TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subunit-like
protein, partial (20%)
Length = 979
Score = 42.7 bits (99), Expect = 9e-06
Identities = 22/74 (29%), Positives = 34/74 (45%)
Frame = +1
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
L VSGG + ++VW+ + L GH D IR + + +S S D IR+W+
Sbjct: 433 LFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQS 612
Query: 67 QRCLHSYAVHTDSV 80
+ C+ H V
Sbjct: 613 RTCISVLTGHNHYV 654
Score = 38.5 bits (88), Expect = 2e-04
Identities = 18/65 (27%), Positives = 31/65 (47%)
Frame = +1
Query: 16 VVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAV 75
V+++WD R G+ K H +R + + +SG D I++W+ RCL +
Sbjct: 334 VIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRCLFTLLG 513
Query: 76 HTDSV 80
H D +
Sbjct: 514 HLDYI 528
Score = 37.7 bits (86), Expect = 3e-04
Identities = 20/81 (24%), Positives = 37/81 (44%)
Frame = +1
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQ 67
+VS ++ +R+W+ +S + L GH + L +S S D +R+WD+G
Sbjct: 562 IVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDIGSL 741
Query: 68 RCLHSYAVHTDSVWALASTST 88
+ A D + L+ +T
Sbjct: 742 K--RKNASPADDILRLSQMNT 798
Score = 25.4 bits (54), Expect = 1.5
Identities = 21/70 (30%), Positives = 33/70 (47%)
Frame = +1
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
L+VS ++ VRVWD GS K D+I L +T F G D++++ G
Sbjct: 685 LVVSASLDQTVRVWD--IGSLKRKNASPADDILRLSQMNTDLF---GGVDAVVKYVLEGH 849
Query: 67 QRCLHSYAVH 76
R ++ + H
Sbjct: 850 DRGVNWASFH 879
>AV409882
Length = 422
Score = 41.6 bits (96), Expect = 2e-05
Identities = 19/56 (33%), Positives = 32/56 (56%)
Frame = +3
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWD 63
+V+G + +RV++ + K + HTD IR + + T + LS S D +I+LWD
Sbjct: 141 VVAGADDMFIRVYNYNTMDKVKVFEAHTDYIRCVAVHPTLPYVLSSSDDMLIKLWD 308
>TC9693 similar to UP|COPP_MOUSE (O55029) Coatomer beta' subunit
(Beta'-coat protein) (Beta'-COP) (p102), partial (20%)
Length = 743
Score = 41.6 bits (96), Expect = 2e-05
Identities = 19/56 (33%), Positives = 32/56 (56%)
Frame = +1
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWD 63
+V+G + +RV++ + K + HTD IR + + T + LS S D +I+LWD
Sbjct: 421 VVAGADDMFIRVYNYNTMDKVKVFEAHTDYIRCVAVHPTLPYVLSSSDDMLIKLWD 588
>TC13023 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragment),
partial (18%)
Length = 591
Score = 41.2 bits (95), Expect = 3e-05
Identities = 21/85 (24%), Positives = 39/85 (45%)
Frame = +1
Query: 17 VRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAVH 76
+++ D + S L+G ++++ AL L S IR+WDL +C+ S+ H
Sbjct: 229 IKIVDSANASIRSTLQGDSESVTALALSPDDNLLFSSGHSRQIRVWDLSTLKCVRSWKGH 408
Query: 77 TDSVWALASTSTFSHVYSGGRDFSV 101
V ++ + + +GG D V
Sbjct: 409 EGPVMCMSCHPSGGLLATGGADRKV 483
Score = 40.8 bits (94), Expect = 3e-05
Identities = 19/70 (27%), Positives = 31/70 (44%)
Frame = +1
Query: 7 LLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQ 66
LL S G + +RVWD + KGH + + +G +G +D + +WD+
Sbjct: 325 LLFSSGHSRQIRVWDLSTLKCVRSWKGHEGPVMCMSCHPSGGLLATGGADRKVLVWDVDG 504
Query: 67 QRCLHSYAVH 76
C H + H
Sbjct: 505 GYCTHFFKGH 534
Score = 30.0 bits (66), Expect = 0.061
Identities = 14/30 (46%), Positives = 17/30 (56%)
Frame = +1
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGH 34
G LL +GG ++ V VWD G T KGH
Sbjct: 445 GGLLATGGADRKVLVWDVDGGYCTHFFKGH 534
>TC19190 weakly similar to PIR|T46032|T46032 WD-40 repeat regulatory
protein tup1 homolog - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (18%)
Length = 511
Score = 41.2 bits (95), Expect = 3e-05
Identities = 19/54 (35%), Positives = 30/54 (55%)
Frame = +3
Query: 45 STGRFCLSGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALASTSTFSHVYSGGRD 98
+ GR+ +SGS D + LWDL Q+ + HTD+V ++ T + + S G D
Sbjct: 9 TNGRYIVSGSEDRCVYLWDLQQKNMIQKLEGHTDTVISVTCHPTENKIASAGLD 170
Score = 36.6 bits (83), Expect = 7e-04
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 2/60 (3%)
Frame = +3
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSG--SSDSMIRLW 62
G +VSG ++ V +WD + + KL+GHTD + ++ T S D +R+W
Sbjct: 15 GRYIVSGSEDRCVYLWDLQQKNMIQKLEGHTDTVISVTCHPTENKIASAGLDGDRTVRVW 194
>TC16458 similar to UP|O48679 (O48679) F3I6.5 protein, partial (20%)
Length = 539
Score = 40.8 bits (94), Expect = 3e-05
Identities = 23/71 (32%), Positives = 37/71 (51%), Gaps = 6/71 (8%)
Frame = +3
Query: 1 MNEGGTLLVSGGTEKVVRVWDPRSGS------KTMKLKGHTDNIRALLLDSTGRFCLSGS 54
+NE G++L SG ++ + VW+ G L+GHT I L++ G SGS
Sbjct: 81 LNEDGSVLYSGACDRSILVWERDGGDGGGEMVVVGALRGHTKAILCLVV--VGDLVFSGS 254
Query: 55 SDSMIRLWDLG 65
+D+ +R+W G
Sbjct: 255 ADNSVRVWRRG 287
>AV413963
Length = 313
Score = 40.8 bits (94), Expect = 3e-05
Identities = 22/64 (34%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Frame = +2
Query: 5 GTLLVSGGTEKVVRVWDPRSGS----KTMKLKGHTDNIRALLLD-STGRFCLSGSSDSMI 59
GT L SG ++ R+W K ++LKGHTD++ L D + S D +
Sbjct: 113 GTKLASGSVDQTARIWHVEQHGHGKVKDIELKGHTDSVDQLCWDPKHADLIATASGDKTV 292
Query: 60 RLWD 63
RLWD
Sbjct: 293 RLWD 304
Score = 24.6 bits (52), Expect = 2.6
Identities = 17/67 (25%), Positives = 26/67 (38%), Gaps = 8/67 (11%)
Frame = +2
Query: 22 PRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCLHSYAV------ 75
P + + GH + ++ + G SGS D R+W + Q H +
Sbjct: 38 PFKNLHSREYSGHKKKVHSVAWNCIGTKLASGSVDQTARIWHVEQ----HGHGKVKDIEL 205
Query: 76 --HTDSV 80
HTDSV
Sbjct: 206 KGHTDSV 226
>TC12572 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (51%)
Length = 770
Score = 38.9 bits (89), Expect = 1e-04
Identities = 24/99 (24%), Positives = 50/99 (50%), Gaps = 5/99 (5%)
Frame = +3
Query: 8 LVSGGTEKVVRVWDPRSGSKTMKLKGHTDN---IRALLLDSTGRFCLSGSSDSMIRLWDL 64
++ G + +R+W+ +G GH ++ I + + G++ + GS D I LW+L
Sbjct: 213 ILVGTLDNNLRLWNYSTGRFLKTYTGHVNSKYCISSTFSTTNGKYVVGGSEDHGIYLWEL 392
Query: 65 GQQRCLHSYAVHTDSVWALASTSTFSHVYSG--GRDFSV 101
++ + H+D+V +++ T + + SG G D +V
Sbjct: 393 QTRKIVQKLEGHSDTVVSVSCHPTENMIASGALGNDKTV 509
Score = 32.3 bits (72), Expect = 0.012
Identities = 14/48 (29%), Positives = 23/48 (47%)
Frame = +3
Query: 23 RSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDLGQQRCL 70
+SG L H+D + A+ + G +S S D + R+WD C+
Sbjct: 3 KSGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDASTGHCM 146
Score = 32.3 bits (72), Expect = 0.012
Identities = 14/60 (23%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Frame = +3
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGS--SDSMIRLW 62
G +V G + + +W+ ++ KL+GH+D + ++ T SG+ +D +++W
Sbjct: 339 GKYVVGGSEDHGIYLWELQTRKIVQKLEGHSDTVVSVSCHPTENMIASGALGNDKTVKIW 518
>TC17738 similar to UP|O48679 (O48679) F3I6.5 protein, partial (8%)
Length = 599
Score = 38.5 bits (88), Expect = 2e-04
Identities = 27/90 (30%), Positives = 43/90 (47%), Gaps = 7/90 (7%)
Frame = +2
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTM----KLKGHTDNIRALLLDSTGRFCLSGSSDSMIR 60
G++L SG ++ + VW+ + M L+GH I L+ + F LSGS+D +R
Sbjct: 2 GSVLFSGACDRSILVWEREDSANHMVVSGALRGHEKAILCLI--NVSDFLLSGSADRTVR 175
Query: 61 LWDLGQQR---CLHSYAVHTDSVWALASTS 87
+W G CL H V +LA+ +
Sbjct: 176VWKRGFDGPFCCLAVLDGHRKPVKSLAAVA 265
>TC9007 similar to UP|Q9SSS7 (Q9SSS7) F6D8.2 protein (At1g52730), partial
(23%)
Length = 647
Score = 38.5 bits (88), Expect = 2e-04
Identities = 18/61 (29%), Positives = 29/61 (47%)
Frame = +3
Query: 5 GTLLVSGGTEKVVRVWDPRSGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
G ++GG + + V+D +G++ KGH + + G SGS D IR+W
Sbjct: 57 GNKFIAGGEDMWIHVFDFHTGNEIACNKGHHGPVHCVRFSPGGESYASGSEDGTIRIWQT 236
Query: 65 G 65
G
Sbjct: 237 G 239
>TC15924 weakly similar to GB|BAC21577.1|24060129|AP005438 G-protein beta
family-like protein {Oryza sativa (japonica
cultivar-group);}, partial (11%)
Length = 1262
Score = 37.4 bits (85), Expect = 4e-04
Identities = 20/64 (31%), Positives = 34/64 (52%), Gaps = 2/64 (3%)
Frame = +1
Query: 7 LLVSGGTEKVVRVWDPR--SGSKTMKLKGHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
LL SG ++ ++VWD R S S +K H ++ + LSGS+D I++W +
Sbjct: 115 LLFSGYSDGSIKVWDIRGHSASLVWDIKEHKKSVTCFSVHEPSDSLLSGSTDKTIKVWKM 294
Query: 65 GQQR 68
Q++
Sbjct: 295 IQRK 306
Score = 22.7 bits (47), Expect = 9.7
Identities = 11/32 (34%), Positives = 15/32 (46%)
Frame = +1
Query: 33 GHTDNIRALLLDSTGRFCLSGSSDSMIRLWDL 64
GH +I L SG SD I++WD+
Sbjct: 67 GHNISIAVCSLIYYKGLLFSGYSDGSIKVWDI 162
>TC18293 similar to UP|PWP1_HUMAN (Q13610) Periodic tryptophan protein 1
homolog (Keratinocyte protein IEF SSP 9502), partial
(7%)
Length = 804
Score = 36.6 bits (83), Expect = 7e-04
Identities = 29/107 (27%), Positives = 47/107 (43%), Gaps = 25/107 (23%)
Frame = +2
Query: 3 EGGTLLVSGGTEKVVRVWD---------------------PRSGSK-TMKLK--GHTDNI 38
E G L G E + +WD ++G+K ++K K HTD+
Sbjct: 200 EKGNFLAVGSMEPSIEIWDLDVLDEVQPCVVLGGIEKKKKGKAGTKKSIKYKDGSHTDSX 379
Query: 39 RALLLDSTGRFCL-SGSSDSMIRLWDLGQQRCLHSYAVHTDSVWALA 84
L + R L S S+D +++WD+ ++C + HTD V A+A
Sbjct: 380 LGLAWNKEYRNILASASADKRVKIWDVVAEKCDITMEHHTDKVQAVA 520
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,068,767
Number of Sequences: 28460
Number of extensions: 28092
Number of successful extensions: 253
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 210
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 247
length of query: 108
length of database: 4,897,600
effective HSP length: 84
effective length of query: 24
effective length of database: 2,506,960
effective search space: 60167040
effective search space used: 60167040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 47 (22.7 bits)
Lotus: description of TM0062.3


