

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
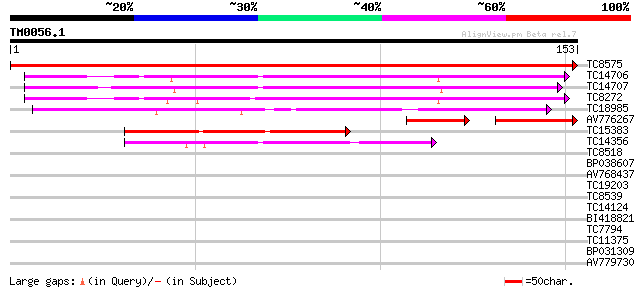
Query= TM0056.1
(153 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8575 weakly similar to UP|Q8L9L8 (Q8L9L8) Pollen allergen-like... 305 2e-84
TC14706 weakly similar to UP|AAQ72568 (AAQ72568) Ripening-relate... 76 3e-15
TC14707 weakly similar to UP|AAQ72568 (AAQ72568) Ripening-relate... 65 7e-12
TC8272 similar to UP|AAQ72568 (AAQ72568) Ripening-related protei... 62 6e-11
TC18985 weakly similar to UP|Q9LQT7 (Q9LQT7) F10B6.35, partial (7%) 53 3e-08
AV776267 35 2e-07
TC15383 43 3e-05
TC14356 41 8e-05
TC8518 similar to UP|Q84MC7 (Q84MC7) At1g01360, partial (84%) 32 0.049
BP038607 30 0.24
AV768437 28 0.70
TC19203 similar to GB|AAO64824.1|29028890|BT005889 At1g20410 {Ar... 27 1.2
TC8539 similar to UP|Q94KE3 (Q94KE3) AT3g52990/F8J2_160 (Pyruvat... 27 1.6
TC14124 UP|Q8H945 (Q8H945) Phosphoenolpyruvate carboxylase , co... 27 1.6
BI418821 27 2.0
TC7794 homologue to UP|Q9M600 (Q9M600) Chlorophyll a/b binding p... 26 2.7
TC11375 similar to UP|Q8L7B2 (Q8L7B2) Serine carboxypeptidase 1-... 26 3.5
BP031309 25 7.7
AV779730 25 7.7
>TC8575 weakly similar to UP|Q8L9L8 (Q8L9L8) Pollen allergen-like protein,
partial (79%)
Length = 762
Score = 305 bits (781), Expect = 2e-84
Identities = 153/153 (100%), Positives = 153/153 (100%)
Frame = +3
Query: 1 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 60
MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY
Sbjct: 30 MGTRGKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITY 209
Query: 61 AEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEVKWSAE 120
AEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEVKWSAE
Sbjct: 210 AEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEVKWSAE 389
Query: 121 YEKASTDIPDPSVVKDFAVKNFLEVDEYVLQQA 153
YEKASTDIPDPSVVKDFAVKNFLEVDEYVLQQA
Sbjct: 390 YEKASTDIPDPSVVKDFAVKNFLEVDEYVLQQA 488
>TC14706 weakly similar to UP|AAQ72568 (AAQ72568) Ripening-related protein,
partial (39%)
Length = 701
Score = 75.9 bits (185), Expect = 3e-15
Identities = 51/155 (32%), Positives = 81/155 (51%), Gaps = 8/155 (5%)
Frame = +3
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSI-------EILEGDGKAAGSVRH 57
GKL +I+LKS A K++ +F K HD + I ++ EGD GSV+
Sbjct: 45 GKLITEIELKSPAVKFY-------NLFAKQL-HDAQKISDSVHETKVHEGDWHGIGSVKQ 200
Query: 58 ITYAEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSE-VK 116
TY ++ + E+I+A D++ KT Y + G++ +YKKF VI +G + VK
Sbjct: 201 WTYVVDGKVI-TCLERIEAIDEQNKTNKYTLFGGDVSSHYKKFTLIFQVIENNDGHDAVK 377
Query: 117 WSAEYEKASTDIPDPSVVKDFAVKNFLEVDEYVLQ 151
W+ EYEK DI P+ D+ K +VD ++++
Sbjct: 378 WTVEYEKLREDIEPPNGYMDYFNKLTRDVDAHLIK 482
>TC14707 weakly similar to UP|AAQ72568 (AAQ72568) Ripening-related protein,
partial (63%)
Length = 731
Score = 64.7 bits (156), Expect = 7e-12
Identities = 44/148 (29%), Positives = 74/148 (49%), Gaps = 3/148 (2%)
Frame = +1
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIE--ILEGDGKAAGSVRHITYAE 62
GKL ++ +K+ ADK++ TT H K E + EGD GSV+H TY
Sbjct: 67 GKLITEVGVKTPADKFYNLF---TTQLHDVQKHAEKVHETKVHEGDWHGIGSVKHWTYVV 237
Query: 63 GSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEV-KWSAEY 121
++ + E+++A D++ KT Y + G++ +YK F + VI + V +W+ EY
Sbjct: 238 DGKVI-TCLERVEAIDEQNKTNKYTLFGGDIDPHYKNFTLILQVIDNKDSHAVARWTIEY 414
Query: 122 EKASTDIPDPSVVKDFAVKNFLEVDEYV 149
EK DI P+ ++ + +VD +
Sbjct: 415 EKLHEDIEPPNGYMEYFNQLTRDVDNQI 498
>TC8272 similar to UP|AAQ72568 (AAQ72568) Ripening-related protein, partial
(64%)
Length = 757
Score = 61.6 bits (148), Expect = 6e-11
Identities = 44/157 (28%), Positives = 81/157 (51%), Gaps = 10/157 (6%)
Frame = +3
Query: 5 GKLEVDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKS-------IEILEGDG--KAAGSV 55
GKL +I +K+ A K++ +F K HD ++ ++ EG+ + ++
Sbjct: 87 GKLSTEIGIKAPAAKFY-------DLFAKQL-HDVQNHCERVHHTKLHEGEDWHEVGSAL 242
Query: 56 RHITYAEGSPLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSE- 114
+H TY V + E+I++ D+E KT+ Y++ DG++ Q+YK F T+ VI +G +
Sbjct: 243 KHWTYVIDGE-VHTCQERIESVDEENKTIKYSLFDGDIGQHYKTFLLTLQVIENKDGHDT 419
Query: 115 VKWSAEYEKASTDIPDPSVVKDFAVKNFLEVDEYVLQ 151
VKW+ EYE + P ++ K ++D +L+
Sbjct: 420 VKWTVEYEMHHEEKEPPHGWMEYVHKCTRDIDANLLK 530
>TC18985 weakly similar to UP|Q9LQT7 (Q9LQT7) F10B6.35, partial (7%)
Length = 552
Score = 52.8 bits (125), Expect = 3e-08
Identities = 40/142 (28%), Positives = 65/142 (45%), Gaps = 2/142 (1%)
Frame = +3
Query: 7 LEVDIDLKSNADKYWLTLRDSTTIFPKAFPHD-YKSIEILEGDGKAAGSVRHITYA-EGS 64
LE + LKS +K + + PK + +EI +GD + GSV+ + EG
Sbjct: 69 LETAVQLKSCPEKLYNLFKSQNQHIPKKTQSEKLHGVEIHKGDWETPGSVKIWKFCIEGK 248
Query: 65 PLVKSSTEKIDAADDEKKTVSYAVIDGELLQYYKKFKGTISVIAVGEGSEVKWSAEYEKA 124
V E+I+ DD +T++Y + G +L+ Y+ +K + V E +K EYEK
Sbjct: 249 EEVFK--ERIEL-DDLNRTITYVAVGGNVLELYRTYKTIVKV----ENGILKLRIEYEKL 407
Query: 125 STDIPDPSVVKDFAVKNFLEVD 146
D P P + F ++D
Sbjct: 408 HEDTPPPKKYQQFVSNIVRDID 473
>AV776267
Length = 382
Score = 34.7 bits (78), Expect(2) = 2e-07
Identities = 15/22 (68%), Positives = 19/22 (86%)
Frame = -2
Query: 132 SVVKDFAVKNFLEVDEYVLQQA 153
S+V+DFAV NFLE+DEY+L A
Sbjct: 306 SLVQDFAVTNFLELDEYILPPA 241
Score = 34.7 bits (78), Expect(2) = 2e-07
Identities = 15/17 (88%), Positives = 16/17 (93%)
Frame = -3
Query: 108 AVGEGSEVKWSAEYEKA 124
A+GEGSEVKWSA YEKA
Sbjct: 380 ALGEGSEVKWSAGYEKA 330
>TC15383
Length = 729
Score = 42.7 bits (99), Expect = 3e-05
Identities = 20/61 (32%), Positives = 38/61 (61%)
Frame = +3
Query: 32 PKAFPHDYKSIEILEGDGKAAGSVRHITYAEGSPLVKSSTEKIDAADDEKKTVSYAVIDG 91
P+A+ H KS ++ GDG G++R + G P +S TE+++ D+E+ +S++V+ G
Sbjct: 519 PQAYKHFVKSCHVIGGDGNV-GTLREVHVISGLPAGRS-TERLEILDEERHVLSFSVVGG 692
Query: 92 E 92
+
Sbjct: 693 D 695
>TC14356
Length = 1330
Score = 41.2 bits (95), Expect = 8e-05
Identities = 26/87 (29%), Positives = 45/87 (50%), Gaps = 3/87 (3%)
Frame = +1
Query: 32 PKAFPHDYKSIEILE--GDGKA-AGSVRHITYAEGSPLVKSSTEKIDAADDEKKTVSYAV 88
P+A+ H K+ ++ GDG G++R + G P SSTE+++ DDE +S++V
Sbjct: 448 PQAYKHFVKTCRVVSVSGDGGIHVGALREVRVVSGLPAA-SSTERLEILDDEGHVMSFSV 624
Query: 89 IDGELLQYYKKFKGTISVIAVGEGSEV 115
+ G+ + +K +V G G V
Sbjct: 625 VGGD--NRLRNYKSVTTVHGDGNGGTV 699
>TC8518 similar to UP|Q84MC7 (Q84MC7) At1g01360, partial (84%)
Length = 665
Score = 32.0 bits (71), Expect = 0.049
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Frame = +2
Query: 36 PHDYKSIE---ILEGDGKAAGSVRHITYAEGSPLVKSSTEKIDAADDEKKTVSYAVIDGE 92
P YK I++GD GSVR + G P +STE+++ DDE+ + ++ G+
Sbjct: 305 PQKYKPFVSRCIMQGD-LGIGSVREVNVKSGLPAT-TSTERLEQLDDEEHILGIRIVGGD 478
>BP038607
Length = 583
Score = 29.6 bits (65), Expect = 0.24
Identities = 18/65 (27%), Positives = 27/65 (40%)
Frame = +2
Query: 31 FPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGSPLVKSSTEKIDAADDEKKTVSYAVID 90
F K FP + K + I D + H ++A P V + DD +SY +D
Sbjct: 398 FTKNFPPELKGLAINNSDAIRSA---HNSFARPEPFVPEEQKTAGKDDDVYHFISYIPVD 568
Query: 91 GELLQ 95
G L +
Sbjct: 569 GVLYE 583
>AV768437
Length = 393
Score = 28.1 bits (61), Expect = 0.70
Identities = 14/29 (48%), Positives = 17/29 (58%), Gaps = 3/29 (10%)
Frame = +3
Query: 92 ELLQYYKKFKGTISVIAVG---EGSEVKW 117
++L Y F+GTIS I V EG EV W
Sbjct: 252 QILAYLNYFRGTISTIQVSQG*EGQEVTW 338
>TC19203 similar to GB|AAO64824.1|29028890|BT005889 At1g20410 {Arabidopsis
thaliana;}, partial (23%)
Length = 526
Score = 27.3 bits (59), Expect = 1.2
Identities = 21/89 (23%), Positives = 34/89 (37%)
Frame = -1
Query: 9 VDIDLKSNADKYWLTLRDSTTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGSPLVK 68
V ID K N + S+ + P DY + L G + LVK
Sbjct: 250 VYIDAKENIGNFQQLSPFSSNV*SFHAPKDYDGVPPLWGSSAKLSN----------RLVK 101
Query: 69 SSTEKIDAADDEKKTVSYAVIDGELLQYY 97
+ E + ++DEK T+ ++ LL +
Sbjct: 100 TRIENLHPSEDEKSTLKQGMVSASLLHLH 14
>TC8539 similar to UP|Q94KE3 (Q94KE3) AT3g52990/F8J2_160 (Pyruvate
kinase-like protein), partial (81%)
Length = 1430
Score = 26.9 bits (58), Expect = 1.6
Identities = 26/86 (30%), Positives = 40/86 (46%), Gaps = 3/86 (3%)
Frame = +2
Query: 29 TIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGSPLVKSST-EKIDAADDEKKTVSYA 87
T+ PK+ +SIE + G KA SV ++ G+P T E + AA K +
Sbjct: 245 TLGPKS-----RSIEAISGCLKAGMSVARFDFSWGNPEYHQETLENLKAAIKGSKKLCAV 409
Query: 88 VID--GELLQYYKKFKGTISVIAVGE 111
++D G +Q K + IS+ A E
Sbjct: 410 MLDTVGAEMQVVNKAETAISLQADAE 487
>TC14124 UP|Q8H945 (Q8H945) Phosphoenolpyruvate carboxylase , complete
Length = 3367
Score = 26.9 bits (58), Expect = 1.6
Identities = 10/17 (58%), Positives = 13/17 (75%)
Frame = +1
Query: 19 KYWLTLRDSTTIFPKAF 35
K+WL LRDST++F F
Sbjct: 3019 KHWLNLRDSTSVFCSLF 3069
>BI418821
Length = 614
Score = 26.6 bits (57), Expect = 2.0
Identities = 24/87 (27%), Positives = 33/87 (37%)
Frame = -2
Query: 27 STTIFPKAFPHDYKSIEILEGDGKAAGSVRHITYAEGSPLVKSSTEKIDAADDEKKTVSY 86
+ I P A P S+ + AA + +T P +S+ I A+ E V
Sbjct: 454 TVVIAPVAIPSQMPSVATVVATTTAAAASSTVTPLAAPPSAESARGGILASGLESGAVGA 275
Query: 87 AVIDGELLQYYKKFKGTISVIAVGEGS 113
IDG L G + IA EGS
Sbjct: 274 GDIDGFGLVVVAGGDGELDEIAFEEGS 194
>TC7794 homologue to UP|Q9M600 (Q9M600) Chlorophyll a/b binding protein
precursor, partial (97%)
Length = 2581
Score = 26.2 bits (56), Expect = 2.7
Identities = 15/46 (32%), Positives = 23/46 (49%)
Frame = -2
Query: 88 VIDGELLQYYKKFKGTISVIAVGEGSEVKWSAEYEKASTDIPDPSV 133
+I GE+L K K I++++ S+V W + S DPSV
Sbjct: 1854 LIFGEVLWLLAKTKWVITIVSGDFSSQVLWDLRSVQESIRQEDPSV 1717
>TC11375 similar to UP|Q8L7B2 (Q8L7B2) Serine carboxypeptidase 1-like
protein (Serine carboxypeptidase 1 precursor-like
protein), partial (35%)
Length = 874
Score = 25.8 bits (55), Expect = 3.5
Identities = 10/36 (27%), Positives = 24/36 (65%)
Frame = -2
Query: 106 VIAVGEGSEVKWSAEYEKASTDIPDPSVVKDFAVKN 141
++ V + S + WS++ ++T++P PS+ DF++ +
Sbjct: 393 MVHVHKQSHLSWSSQVHHSATELPGPSL--DFSLNS 292
>BP031309
Length = 422
Score = 24.6 bits (52), Expect = 7.7
Identities = 11/23 (47%), Positives = 15/23 (64%)
Frame = +2
Query: 95 QYYKKFKGTISVIAVGEGSEVKW 117
Q + + GTI+ I GEGS+ KW
Sbjct: 293 QLFLQRLGTIAHIYTGEGSKQKW 361
Score = 24.6 bits (52), Expect = 7.7
Identities = 11/23 (47%), Positives = 15/23 (64%)
Frame = +1
Query: 95 QYYKKFKGTISVIAVGEGSEVKW 117
Q + + GTI+ I GEGS+ KW
Sbjct: 220 QLFLQRLGTIAHIYTGEGSKQKW 288
>AV779730
Length = 481
Score = 24.6 bits (52), Expect = 7.7
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = -2
Query: 21 WLTLRDSTTIFPKAF 35
WL+LRDST++F F
Sbjct: 213 WLSLRDSTSVFCSLF 169
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,994,991
Number of Sequences: 28460
Number of extensions: 18745
Number of successful extensions: 82
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 79
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 79
length of query: 153
length of database: 4,897,600
effective HSP length: 82
effective length of query: 71
effective length of database: 2,563,880
effective search space: 182035480
effective search space used: 182035480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0056.1


