

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.3
(355 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
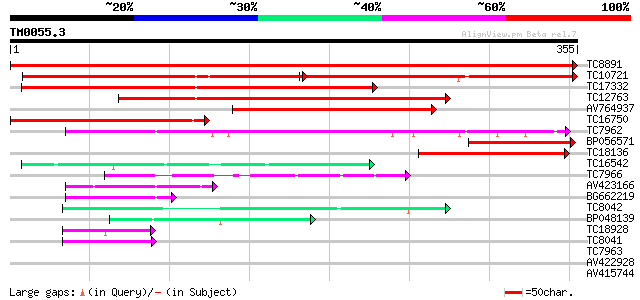
Score E
Sequences producing significant alignments: (bits) Value
TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Ara... 725 0.0
TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehyd... 211 2e-98
TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehyd... 246 3e-66
TC12763 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 179 5e-46
AV764937 142 9e-35
TC16750 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 127 2e-30
TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 83 8e-17
BP056571 77 3e-15
TC18136 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 68 2e-12
TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, pa... 63 9e-11
TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, par... 60 6e-10
AV423166 53 9e-08
BG662219 52 2e-07
TC8042 homologue to UP|P93619 (P93619) Probable NADP-dependent o... 48 3e-06
BP048139 45 2e-05
TC18928 similar to UP|Q43677 (Q43677) Auxin-induced protein, par... 40 5e-04
TC8041 similar to UP|P93619 (P93619) Probable NADP-dependent oxi... 40 5e-04
TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 39 0.002
AV422928 38 0.003
AV415744 31 0.28
>TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , complete
Length = 1636
Score = 725 bits (1872), Expect = 0.0
Identities = 355/355 (100%), Positives = 355/355 (100%)
Frame = +1
Query: 1 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 60
MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN
Sbjct: 94 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 273
Query: 61 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 120
SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF
Sbjct: 274 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 453
Query: 121 TFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQ 180
TFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQ
Sbjct: 454 TFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQ 633
Query: 181 PGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMR 240
PGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMR
Sbjct: 634 PGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMR 813
Query: 241 ALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPGEIKIHPGLLIMGSRTVSGSGV 300
ALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPGEIKIHPGLLIMGSRTVSGSGV
Sbjct: 814 ALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPGEIKIHPGLLIMGSRTVSGSGV 993
Query: 301 GGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSLN 355
GGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSLN
Sbjct: 994 GGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSLN 1158
>TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , complete
Length = 1297
Score = 211 bits (536), Expect(2) = 2e-98
Identities = 101/178 (56%), Positives = 127/178 (70%)
Frame = +3
Query: 9 ECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPG 68
+ GWAA DTSGVLSP+ FSRR GDDDV +KI CGVC++D+ +N G + YP VPG
Sbjct: 108 KAFGWAARDTSGVLSPFHFSRRENGDDDVAIKILFCGVCHSDLHTIKNDWGFTTYPVVPG 287
Query: 69 HEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHD 128
HEI G+VTK G+NV F VGD VGVG V+SC+ CE CD LE++C + V+T+N ++
Sbjct: 288 HEIVGVVTKTGNNVTKFKVGDKVGVGVIVDSCKSCENCDQDLENYCPQ-PVYTYNS-PYN 461
Query: 129 GTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLG 186
GT T GGYS F+VVH+RY P + PL + APLLCAGITVY PM + M +PGK+LG
Sbjct: 462 GTRTNGGYSDFVVVHQRYVLQFPDNLPLDAGAPLLCAGITVYXPMKYYGMTEPGKTLG 635
Score = 165 bits (418), Expect(2) = 2e-98
Identities = 88/177 (49%), Positives = 118/177 (65%), Gaps = 3/177 (1%)
Frame = +1
Query: 182 GKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRA 241
GK LGV GLGGLGH+A+KFGKAFGL VTV STS +K+ EA+ LGAD F+VS+D +M+A
Sbjct: 622 GKHLGVAGLGGLGHVAIKFGKAFGLKVTVISTSPNKEAEAIEKLGADSFLVSTDPAKMKA 801
Query: 242 LAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPG---EIKIHPGLLIMGSRTVSGS 298
++ +I+DT S HS P + LLK G L VG PG E+ + P L+ G + V GS
Sbjct: 802 ALGTIHYIIDTISASHSLIPLLGLLKLNGKLVTVGLPGKPLELPVFP--LVAGRKLVGGS 975
Query: 299 GVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSLN 355
GG KE ++M+EF + I +IE++ I N A++R+ K DVKYRFVID+ S +
Sbjct: 976 HFGGIKETQEMLEFSAKHNIAADIELIKIDEINTAMERLSKADVKYRFVIDVAXSFS 1146
>TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (63%)
Length = 736
Score = 246 bits (629), Expect = 3e-66
Identities = 122/223 (54%), Positives = 156/223 (69%)
Frame = +1
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVP 67
++ GWAA D+SGVLSP+KFSRR G+ DV K+ +CG+C++D+ +N G+SVYP VP
Sbjct: 70 KKAFGWAARDSSGVLSPFKFSRRETGEKDVTFKVLYCGICHSDLHMIKNEWGSSVYPLVP 249
Query: 68 GHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDH 127
GHEIAG+VT+VGS V F VGD VGVG VNSCR C+ C D LE++C K + T+ +
Sbjct: 250 GHEIAGVVTEVGSKVQKFKVGDRVGVGCMVNSCRSCQSCADNLENYCPK-MILTYAAKNV 426
Query: 128 DGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGV 187
DGTIT GGYS +V E + I + PL +AAP LCAGITVYSP+ + +++PG +GV
Sbjct: 427 DGTITYGGYSDLMVSDEDFGIRISDNLPLEAAAPFLCAGITVYSPLKYYGLDKPGLHVGV 606
Query: 188 IGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRF 230
GLGGLGHMAVKF A G VT STS +KK EA+ + AD F
Sbjct: 607 SGLGGLGHMAVKFAIALGAKVTAISTSPNKKTEAIEKV*ADSF 735
>TC12763 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (62%)
Length = 809
Score = 179 bits (455), Expect = 5e-46
Identities = 91/208 (43%), Positives = 131/208 (62%)
Frame = +2
Query: 69 HEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHD 128
HE+ G V +VGS+V F+VG+ VG G V C+ C C +E +C K ++ +N V D
Sbjct: 68 HEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKK-KIWNYNDVYVD 244
Query: 129 GTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVI 188
G T+GG+++ I+V +++ IP+ APLLCA +TVYSP+ + + G G++
Sbjct: 245 GKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGIL 424
Query: 189 GLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDF 248
GLGG+GHM V KA G +VTV S+S KK+EA+ LGAD ++VSSD M+ A SLD+
Sbjct: 425 GLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDY 604
Query: 249 IVDTASGPHSFDPYMALLKSFGVLALVG 276
I+DT H +PY++LLK G L L+G
Sbjct: 605 IIDTVPVGHPLEPYLSLLKLDGKLILMG 688
>AV764937
Length = 392
Score = 142 bits (358), Expect = 9e-35
Identities = 71/129 (55%), Positives = 96/129 (74%), Gaps = 1/129 (0%)
Frame = +1
Query: 140 IVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKM-NQPGKSLGVIGLGGLGHMAV 198
+V RY IP++ P+ +AAPLLCAGITV+SP+ H + + PGK +GV+GLGGLGH+AV
Sbjct: 4 MVADYRYVVHIPENLPMDAAAPLLCAGITVFSPLKDHDLVSSPGKKIGVVGLGGLGHIAV 183
Query: 199 KFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASGPHS 258
KFGKAFG +VTV STS SK+ EA LGAD F++S++Q+ +++ +SLDF+ DT S HS
Sbjct: 184 KFGKAFGHHVTVISTSPSKEAEAKQRLGADDFILSTNQQHLQSARRSLDFVSDTVSAEHS 363
Query: 259 FDPYMALLK 267
P + LLK
Sbjct: 364 LLPLLELLK 390
>TC16750 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (34%)
Length = 442
Score = 127 bits (320), Expect = 2e-30
Identities = 60/125 (48%), Positives = 79/125 (63%)
Frame = +1
Query: 1 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 60
M S A +GWAA D SG+LSPY F+ R G DDVY+K+ +CGVC+ DV +N LG
Sbjct: 64 MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM 243
Query: 61 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 120
S YP VPGHE+ G V +VGS+V F+VG+ VG G V C+ C C +E +C K ++
Sbjct: 244 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYC-KKKIW 420
Query: 121 TFNGV 125
+N V
Sbjct: 421 NYNDV 435
>TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , complete
Length = 1489
Score = 82.8 bits (203), Expect = 8e-17
Identities = 80/346 (23%), Positives = 141/346 (40%), Gaps = 30/346 (8%)
Frame = +1
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI + +C+ DV + ++P + GHE GIV VG V GDH +
Sbjct: 220 EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPGDHA-LPV 396
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGV---DHDGTITKGG-----------YSKFIV 141
+ C +C C + C + T GV D+ + G +S++ V
Sbjct: 397 FTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSRFSIKGKPIYHFVGTSTFSEYTV 576
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
+H I + PL L C T + + +PG S+ + GLG +G A +
Sbjct: 577 LHAGCVAKINPAAPLDKVCILSCGICTGFGATVNVAKPKPGSSVAIFGLGAVGLAAAEGA 756
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE--MRALAKSLDFIVD-----TAS 254
+ G + + +S + E G + FV D ++ +A+ + VD T S
Sbjct: 757 RVSGASRIIGVDLVSSRFEGAKKFGVNEFVNPKDHDKPVQEVIAEMTNGGVDRAVECTGS 936
Query: 255 GPHSFDPYMALLKSFGVLALVGFPGE---IKIHPGLLIMGSRTVSGSGVGGTK---EIRD 308
+ + +GV LVG P + + HP + + RT+ G+ G K ++ +
Sbjct: 937 IQAMISAFECVHDGWGVAVLVGVPNQDDAFQTHP-VNFLNERTLKGTFYGNYKPRTDLPN 1113
Query: 309 MIEFCVANEIHPN---IEVVPIQYANEALDRVIKKDVKYRFVIDIE 351
++E + E+ V N+A D ++K + R +I +E
Sbjct: 1114VVEMYMRGELELEKFITHTVSFSEINQAFDYMLKGE-SIRCIIRME 1248
>BP056571
Length = 319
Score = 77.4 bits (189), Expect = 3e-15
Identities = 33/67 (49%), Positives = 51/67 (75%)
Frame = -1
Query: 288 LIMGSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFV 347
L+ G +T++ S +GG KE ++MI+F +++ P IEV+P+ Y N A++R++K DVKYRFV
Sbjct: 301 LLGGRKTIAXSMIGGIKETQEMIDFAAKHDVKPEIEVIPMDYVNTAMERLLKADVKYRFV 122
Query: 348 IDIENSL 354
IDI N+L
Sbjct: 121 IDIGNTL 101
>TC18136 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (28%)
Length = 601
Score = 68.2 bits (165), Expect = 2e-12
Identities = 33/95 (34%), Positives = 59/95 (61%), Gaps = 1/95 (1%)
Frame = +3
Query: 257 HSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSGVGGTKEIRDMIEFCVA 315
H +PY++LLK G L L+G ++ ++++G R+++GS +G KE +M+EF
Sbjct: 9 HPLEPYLSLLKLDGKLILMGVIYTPLQFITPMVMLGRRSITGSFIGSIKETEEMVEFWKE 188
Query: 316 NEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDI 350
+ IE+V + Y ++ +R+ K DV+YRFV+D+
Sbjct: 189 KGLTSMIEIVNMDYIHKPFERLEKNDVRYRFVVDV 293
>TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, partial (60%)
Length = 724
Score = 62.8 bits (151), Expect = 9e-11
Identities = 57/224 (25%), Positives = 88/224 (38%), Gaps = 3/224 (1%)
Frame = +1
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVY---P 64
E W + + P+K +G DV V++ G+C +DV + + P
Sbjct: 88 ENMAAWLVAINTLKIQPFKLP--TLGPHDVRVRMKAVGICGSDVHYLKKLRCADFIVKEP 261
Query: 65 CVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNG 124
V GHE AGI+ VG+ V GD V + + SC C+ C G + C F
Sbjct: 262 MVIGHECAGIIEAVGAEVKSLVPGDRVAIEPGI-SCWRCDHCKLGSYNLCPDMKFFATPP 438
Query: 125 VDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKS 184
V G + IV CF +P + L A +C ++V R P +
Sbjct: 439 V-------HGSLANQIVHPADLCFKLPDNVSLEEGA--MCEPLSVGVHACRRANIGPETN 591
Query: 185 LGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGAD 228
+ ++G G +G + + +AFG V + LGAD
Sbjct: 592 VLIMGAGPIGLVTMLAARAFGAPRIVIVDVDDHRLSVAKTLGAD 723
>TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, partial (98%)
Length = 1908
Score = 60.1 bits (144), Expect = 6e-10
Identities = 58/194 (29%), Positives = 89/194 (44%), Gaps = 2/194 (1%)
Frame = +2
Query: 60 NSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSV 119
+S P PG+++AG+V KVGS V F VGD V + ++ LE+ V GS+
Sbjct: 902 DSPLPTAPGYDVAGVVVKVGSEVKKFKVGDEV----------YGDINENALEYPKVVGSL 1051
Query: 120 FTFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMN 179
+ V+ ++ H+ PK+ AA L A T Y + R +
Sbjct: 1052 AEYTAVEEK-----------LLAHK------PKNLSFVEAASLPLAIETAYEGLERAGFS 1180
Query: 180 QPGKSLGVI-GLGGLGHMAVKFGK-AFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQE 237
GKS+ V+ G GG+G ++ K FG + V +TS + K E L LGAD + +E
Sbjct: 1181 -AGKSILVLGGAGGVGTHVIQLAKHVFGAS-KVAATSSTGKLELLRKLGAD-MPIDYTKE 1351
Query: 238 EMRALAKSLDFIVD 251
+ L + D + D
Sbjct: 1352 NVENLPEKFDVVFD 1393
>AV423166
Length = 452
Score = 52.8 bits (125), Expect = 9e-08
Identities = 29/95 (30%), Positives = 49/95 (51%)
Frame = +2
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V VK+ +C+ D+ + + + ++P GHE GIV VG V GD + + T
Sbjct: 158 EVRVKMLCASICHTDIT-SIHGFPHGIFPLALGHEGVGIVESVGDQVTNLKEGDMI-IPT 331
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGT 130
Y+ C++CE C G + C+K + +G+ D T
Sbjct: 332 YIGECQECENCVSGETNLCMKYPI-ALSGMMPDNT 433
>BG662219
Length = 366
Score = 51.6 bits (122), Expect = 2e-07
Identities = 25/69 (36%), Positives = 36/69 (51%)
Frame = +2
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V VKI + +C+ D ++PC+ GHE AG+V VG V GDHV +
Sbjct: 161 EVRVKILYTALCHTDAYTWSGKDPEGLFPCILGHEAAGVVESVGEGVTEVQPGDHV-IPC 337
Query: 96 YVNSCRDCE 104
Y CR+C+
Sbjct: 338 YQAECRECK 364
>TC8042 homologue to UP|P93619 (P93619) Probable NADP-dependent
oxidoreductase TED2 , complete
Length = 1244
Score = 47.8 bits (112), Expect = 3e-06
Identities = 56/249 (22%), Positives = 93/249 (36%), Gaps = 6/249 (2%)
Frame = +2
Query: 34 DDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGV 93
+++V VK GV + DV + + S P PG E G+VT VG G VGD VG
Sbjct: 197 ENEVRVKNKAIGVNFIDVYFRKGVYKASTSPFTPGMEAVGVVTAVGGGPTGVQVGDLVGY 376
Query: 94 GTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTITKGGYSKFIVVHERYCFLIPKS 153
G Y + ++H IP S
Sbjct: 377 AGQ-----------------------------------PIGSYVEEQILHAGKVVPIPSS 451
Query: 154 YPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGV-IGLGGLGHMAVKFGKAFGLNVTVFS 212
+ AA ++ G+T + R + G ++ V GG+G + ++ A G TV
Sbjct: 452 IDPSVAASVMLKGMTAQFLLRRCFKVERGHTILVHAAAGGVGSLLCQWANALG--ATVIG 625
Query: 213 TSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDF-----IVDTASGPHSFDPYMALLK 267
T ++++ A + V+ +E+ A + +V + G +F+ +A LK
Sbjct: 626 TVSNEEKAAQAKEDGCHHVIIYKEEDFVARVNEITSGNGVEVVYDSVGKDTFEGSLACLK 805
Query: 268 SFGVLALVG 276
G + G
Sbjct: 806 QRGYMVSFG 832
>BP048139
Length = 567
Score = 44.7 bits (104), Expect = 2e-05
Identities = 35/143 (24%), Positives = 53/143 (36%), Gaps = 14/143 (9%)
Frame = -2
Query: 63 YPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTF 122
+P + GHE G+V VG +V + GD V + Y+ C +C C + C
Sbjct: 431 FPRILGHEAVGVVESVGKDVTEVTKGD-VVIPIYLPDCGECTDCKSTKSNRCTNFPFKVS 255
Query: 123 NGVDHDGT--------------ITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGIT 168
+ DGT + +S++ VV I P A L C T
Sbjct: 254 PSMPRDGTTRFTDLNGEIIHHFMFISSFSEYTVVDVANVTKIDPEIPPDRACLLSCGIST 75
Query: 169 VYSPMMRHKMNQPGKSLGVIGLG 191
R +PG ++ + GLG
Sbjct: 74 GVGAAWRTASVEPGSTVAIFGLG 6
>TC18928 similar to UP|Q43677 (Q43677) Auxin-induced protein, partial (53%)
Length = 612
Score = 40.4 bits (93), Expect = 5e-04
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Frame = +1
Query: 34 DDDVYVKISHCGVCYADVAWTRNTL--GNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHV 91
+D V +K++ + D +S P PG+++AG+V KVGS V F VGD V
Sbjct: 172 EDQVLIKVAAASLNPIDYKRMEGAFKASDSPLPTAPGYDVAGVVVKVGSEVKKFKVGDEV 351
>TC8041 similar to UP|P93619 (P93619) Probable NADP-dependent
oxidoreductase TED2 , partial (31%)
Length = 637
Score = 40.4 bits (93), Expect = 5e-04
Identities = 23/59 (38%), Positives = 29/59 (48%)
Frame = +1
Query: 34 DDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVG 92
+ +V VK GV + DV + + S P PG E G+VT VG G VGD VG
Sbjct: 421 EGEVRVKNKAIGVNFIDVYFRKGVYKASSTPFTPGMEAVGVVTAVGGGPTGVQVGDLVG 597
>TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , partial
(33%)
Length = 694
Score = 38.5 bits (88), Expect = 0.002
Identities = 24/90 (26%), Positives = 38/90 (41%)
Frame = +3
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI + +C+ DV + ++P + GHE GIV VG V DH +
Sbjct: 387 EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPVDH-ALPV 563
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGV 125
+ C + C + C + T GV
Sbjct: 564 FTGECGELPHCKSEESNMCDLLRINTDRGV 653
>AV422928
Length = 399
Score = 37.7 bits (86), Expect = 0.003
Identities = 17/33 (51%), Positives = 23/33 (69%)
Frame = -3
Query: 323 EVVPIQYANEALDRVIKKDVKYRFVIDIENSLN 355
E++ NEA++R+ K DV+YRFVIDI S N
Sbjct: 397 ELITPDRINEAMERLAKNDVRYRFVIDIAGSSN 299
>AV415744
Length = 415
Score = 31.2 bits (69), Expect = 0.28
Identities = 35/114 (30%), Positives = 55/114 (47%), Gaps = 10/114 (8%)
Frame = +1
Query: 182 GKSLGVIGL-GGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRF-------VVS 233
G+ L V+G GG+G AV+ GKA G V + ++K + L G D V
Sbjct: 52 GQVLLVLGAAGGVGLAAVQIGKACGAIVIAVARG-AEKVQLLKSHGVDHVVDLTNENVTE 228
Query: 234 SDQEEMRA-LAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGF-PGEIKIHP 285
S +E ++A K +D + D G + + + LLK + ++GF GEI + P
Sbjct: 229 SVKEFLKARKLKGIDVLYDPVGGKLTKE-CLRLLKWGANILIIGFASGEIPVIP 387
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,287,462
Number of Sequences: 28460
Number of extensions: 91013
Number of successful extensions: 417
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 405
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 406
length of query: 355
length of database: 4,897,600
effective HSP length: 91
effective length of query: 264
effective length of database: 2,307,740
effective search space: 609243360
effective search space used: 609243360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0055.3


