

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.1
(241 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
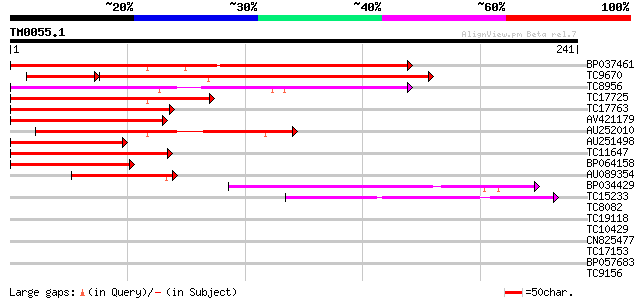
Score E
Sequences producing significant alignments: (bits) Value
BP037461 149 3e-37
TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%) 105 5e-33
TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, pa... 107 1e-24
TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, part... 103 2e-23
TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor,... 102 6e-23
AV421179 92 8e-20
AU252010 84 2e-17
AU251498 84 3e-17
TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, pa... 77 3e-15
BP064158 70 3e-13
AU089354 47 2e-06
BP034429 47 4e-06
TC15233 similar to UP|AAS45681 (AAS45681) AGAMOUS-like protein (... 42 9e-05
TC8082 similar to UP|Q8H9E8 (Q8H9E8) Resistant specific protein-... 30 0.48
TC19118 weakly similar to UP|Q84YE8 (Q84YE8) Expressed protein-l... 29 0.63
TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%) 29 0.63
CN825477 28 1.8
TC17153 UP|AAQ87023 (AAQ87023) VDAC3.1, complete 27 4.1
BP057683 26 5.4
TC9156 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Frag... 26 5.4
>BP037461
Length = 551
Score = 149 bits (377), Expect = 3e-37
Identities = 82/175 (46%), Positives = 116/175 (65%), Gaps = 4/175 (2%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEY-AS 59
MGRG++ ++RI+N +RQVTF+KRRNGLLKKA ELS+LCDAEV LI+FS+ GKLYE+ +S
Sbjct: 8 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 187
Query: 60 TSMKSVIERYNKLK---EEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLS 116
+SM +ERY K E N A EL Q E L+ + + LQ R LMGE L
Sbjct: 188 SSMLKTLERYQKCNYGAPEANVSTREALELSS-QQEYLKLKARYEALQRSQRNLMGEDLG 364
Query: 117 GLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKME 171
L KEL++LE QL+ SLK +R + Q + D++ +L +K +++ + N L +++E
Sbjct: 365 PLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLE 529
>TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%)
Length = 1060
Score = 105 bits (263), Expect(2) = 5e-33
Identities = 57/143 (39%), Positives = 89/143 (61%), Gaps = 1/143 (0%)
Frame = +3
Query: 39 CDAEVGLIVFSSTGKLYEYASTSMKSVIERYNKLKEEHNQLMNPA-SELKFWQTEAASLR 97
CDAEV LIVFSS G+LYEYA+ S+K+ I+RY K + + + + + +F+Q EA LR
Sbjct: 93 CDAEVALIVFSSRGRLYEYANNSVKATIDRYKKACSDSSGAGSASEANAQFYQQEADKLR 272
Query: 98 QQLQYLQECHRQLMGEGLSGLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGN 157
Q+ LQ +RQ+MGE L + KEL+NLE +LE + +R KK+++L EI+ + ++
Sbjct: 273 VQISNLQNNNRQMMGESLGSMNAKELKNLETKLEKGISRIRSKKNELLFAEIEYMQKREI 452
Query: 158 LIHQENAELYKKMELIQKENAEL 180
+H N L K+ ++ + L
Sbjct: 453 DLHNNNQLLRAKIAESERNHPNL 521
Score = 51.2 bits (121), Expect(2) = 5e-33
Identities = 24/31 (77%), Positives = 29/31 (93%)
Frame = +1
Query: 8 IRRIDNSTSRQVTFSKRRNGLLKKAKELSIL 38
I+RI+N+T+RQVTF KRRNGLLKKA ELS+L
Sbjct: 1 IKRIENTTNRQVTFCKRRNGLLKKAYELSVL 93
>TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, partial
(58%)
Length = 1100
Score = 107 bits (268), Expect = 1e-24
Identities = 63/182 (34%), Positives = 102/182 (55%), Gaps = 11/182 (6%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI I+ I+N T+RQVT+SKRRNG+ KKA ELS+LCDA+V LI+FS K++EY S
Sbjct: 137 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYISP 316
Query: 61 SM--KSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQL-------M 111
+ K +I++Y K ++ W++ + + L+ L+E + +L +
Sbjct: 317 GLTTKRIIDQYQK----------TLGDIDLWRSHYEKMLENLKKLKEINNKLRRQIRHRL 466
Query: 112 GEGL--SGLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKK 169
GEGL L ++L+ LE + S+ +R +K ++ +K + Q N L +
Sbjct: 467 GEGLDMDDLSFQQLRKLEEDMVSSIGKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLE 646
Query: 170 ME 171
++
Sbjct: 647 LK 652
>TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, partial (45%)
Length = 970
Score = 103 bits (258), Expect = 2e-23
Identities = 52/88 (59%), Positives = 68/88 (77%), Gaps = 1/88 (1%)
Frame = +3
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEY-AS 59
MGRG++ ++RI+N +RQVTF+KRRNGLLKKA ELS+LCDAEV LIVFS+ GKLYE+ +S
Sbjct: 276 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIVFSTRGKLYEFCSS 455
Query: 60 TSMKSVIERYNKLKEEHNQLMNPASELK 87
+SM +ERY K ++ PA EL+
Sbjct: 456 SSMVKTLERYQKCSYGAVEVNKPAKELR 539
>TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor, partial
(35%)
Length = 737
Score = 102 bits (254), Expect = 6e-23
Identities = 47/70 (67%), Positives = 62/70 (88%)
Frame = +3
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M R KI I++IDN+T+RQVTFSKRR GL KKA+ELS+LCDA+V L+VFSSTGKL+EY++
Sbjct: 501 MAREKIQIKKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALVVFSSTGKLFEYSNL 680
Query: 61 SMKSVIERYN 70
SMK ++ER++
Sbjct: 681 SMKEILERHH 710
>AV421179
Length = 207
Score = 92.0 bits (227), Expect = 8e-20
Identities = 45/67 (67%), Positives = 57/67 (84%)
Frame = +3
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M R +I I++IDN +SRQVTFSKRR GL KKA+ELS LCDA++ LIVFS+T KL+EYAS+
Sbjct: 6 MTRKRIQIKKIDNISSRQVTFSKRRKGLFKKAQELSTLCDADIALIVFSATNKLFEYASS 185
Query: 61 SMKSVIE 67
S++ VIE
Sbjct: 186SIQKVIE 206
>AU252010
Length = 328
Score = 84.3 bits (207), Expect = 2e-17
Identities = 44/120 (36%), Positives = 78/120 (64%), Gaps = 9/120 (7%)
Frame = +1
Query: 12 DNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEY--ASTSMKSVIERY 69
+NS++RQVT+SKR+NG+LKKAKE+++LCDA+V LI+F+++GK+++Y ST++ ++ERY
Sbjct: 1 ENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISPSTTLVDMLERY 180
Query: 70 NKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECH-------RQLMGEGLSGLGVKE 122
+K S + W + +L +++ L++ + R L G+ ++ L KE
Sbjct: 181 HK-----------TSGKRLWDAKHENLNGEIERLKKENDGMQIELRHLKGDDINSLNYKE 327
>AU251498
Length = 342
Score = 83.6 bits (205), Expect = 3e-17
Identities = 41/50 (82%), Positives = 46/50 (92%)
Frame = +3
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSS 50
MGRGKI I+RI+N+T+RQVTF KRRNGLLKKA ELS+LCDAEV LIVFSS
Sbjct: 45 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSS 194
Score = 33.9 bits (76), Expect = 0.026
Identities = 18/48 (37%), Positives = 29/48 (59%), Gaps = 2/48 (4%)
Frame = +1
Query: 33 KELSILCDAEVGLIVFSST--GKLYEYASTSMKSVIERYNKLKEEHNQ 78
K +S C + L SS G+LYEY++ +++S IERY K +H++
Sbjct: 136 KLMSYQCCVMLKLPSLSSPARGRLYEYSNNNIRSTIERYKKACSDHSK 279
>TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, partial (25%)
Length = 843
Score = 76.6 bits (187), Expect = 3e-15
Identities = 35/69 (50%), Positives = 52/69 (74%)
Frame = +1
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
+GR KI ++++ N ++ QVTFSKRR+GL KKA EL ILCD E+ L+VFS K++ +
Sbjct: 85 LGRQKIEMKKMTNESNLQVTFSKRRSGLFKKASELCILCDVEIALVVFSPGEKVFSFGHP 264
Query: 61 SMKSVIERY 69
S+++VI+RY
Sbjct: 265 SVEAVIKRY 291
>BP064158
Length = 394
Score = 70.1 bits (170), Expect = 3e-13
Identities = 32/53 (60%), Positives = 45/53 (84%)
Frame = +3
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGK 53
MGR K+ ++++N+T+RQVTFSKRRNGL+KKA ELS+LC +V LI+FS +G+
Sbjct: 165 MGRVKLQNQKVENTTNRQVTFSKRRNGLIKKAYELSVLCXFDVALIMFSPSGR 323
>AU089354
Length = 178
Score = 47.4 bits (111), Expect = 2e-06
Identities = 22/47 (46%), Positives = 35/47 (73%), Gaps = 2/47 (4%)
Frame = +3
Query: 27 GLLKKAKELSILCDAEVGLIVFSSTGKLYEYASTSMKSV--IERYNK 71
G+LKKA E+++LC A+V LI F+++GK+++Y S+ V +ERY K
Sbjct: 3 GILKKAIEITVLCXAQVSLIXFAASGKMHDYIRPSITXVEMLERYXK 143
>BP034429
Length = 567
Score = 46.6 bits (109), Expect = 4e-06
Identities = 39/139 (28%), Positives = 66/139 (47%), Gaps = 7/139 (5%)
Frame = -1
Query: 94 ASLRQQLQYLQECHRQLMGEGLSGLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELH 153
A L +++ E R++ GE GL +L LE L LK V K++ + DEI +
Sbjct: 546 AELHKEVANRTEQLRRMTGEDFEGLEFDDLLELEKTLPSGLKRVIELKEKRIMDEITAVQ 367
Query: 154 QKGNLIHQENAELYKKMELIQKENAELQKKVYEARSTNEKDAASSPS---CTIRNG---- 206
+KG + +EN L +KM ++ K + + ++ T ++ A+S C+ +G
Sbjct: 366 KKGIQLEEENKLLKQKMAMLCKGKSHF---LLDSNITMQEVASSDSMNNVCSCNSGPSLD 196
Query: 207 YDLQAISLQLSQPQPQFSE 225
D SL+L P P +E
Sbjct: 195 DDSSVTSLKLGLPFPN*NE 139
>TC15233 similar to UP|AAS45681 (AAS45681) AGAMOUS-like protein (Fragment),
partial (40%)
Length = 610
Score = 42.0 bits (97), Expect = 9e-05
Identities = 31/116 (26%), Positives = 55/116 (46%)
Frame = +3
Query: 118 LGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKEN 177
L +KEL+NLE +LE L VR +K + L +++ + ++ I +N Y + ++ + E
Sbjct: 3 LSLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKRE--IELQNHNHYLRAKIAEHER 176
Query: 178 AELQKKVYEARSTNEKDAASSPSCTIRNGYDLQAISLQLSQPQPQFSEPPAKAVKL 233
A+ Q++ + + N + S PS YD L Q+S A+ L
Sbjct: 177 AQQQQQQQQQQPQNLMLSESLPS----QSYDRNFFPANLLGSDNQYSRQDQTALPL 332
>TC8082 similar to UP|Q8H9E8 (Q8H9E8) Resistant specific protein-3, partial
(54%)
Length = 1913
Score = 29.6 bits (65), Expect = 0.48
Identities = 26/94 (27%), Positives = 45/94 (47%), Gaps = 4/94 (4%)
Frame = +1
Query: 68 RYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHR--QLMGEGLSGLGVKELQN 125
+Y K + ++QL P S L + ++ +QY +E QL +S LGVKE+Q+
Sbjct: 640 QYTKESQSNDQL--PISNLGVKEIQSHDNPLWIQYTKESESNDQLP---ISNLGVKEIQS 804
Query: 126 LENQLEMSLKGVRMKKDQILTDE--IKELHQKGN 157
+N L + DQ+ + +KE+ + N
Sbjct: 805 QDNPLWIQYSKESQSNDQLPISKLGVKEIQSQDN 906
Score = 28.5 bits (62), Expect = 1.1
Identities = 27/110 (24%), Positives = 50/110 (44%), Gaps = 2/110 (1%)
Frame = +1
Query: 50 STGKLYEYASTSMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQ 109
S K+ + S+ + +Y K + ++QL P S L + ++ +QY +E +
Sbjct: 214 SKDKISKEISSQDNPLWIQYTKEAQSNDQL--PISNLGVKEIQSQDNPLWIQYTKESQSK 387
Query: 110 LMGEGLSGLGVKELQNLENQLEMSLKGVRMKKDQILTDEI--KELHQKGN 157
+S LGVKE+Q+ +N L + DQ+ + KE+ + N
Sbjct: 388 DQLP-ISNLGVKEIQSHDNPLWIQYTKESQSNDQLPISNLGAKEIQSQDN 534
Score = 27.3 bits (59), Expect = 2.4
Identities = 23/100 (23%), Positives = 44/100 (44%), Gaps = 3/100 (3%)
Frame = +1
Query: 61 SMKSVIERYNKLKEE-HNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLG 119
S+++ ++ Y + K++ ++ + + L T+ A QL +S LG
Sbjct: 181 SIETCVDTYTQSKDKISKEISSQDNPLWIQYTKEAQSNDQLP-------------ISNLG 321
Query: 120 VKELQNLENQLEMSLKGVRMKKDQILTDE--IKELHQKGN 157
VKE+Q+ +N L + KDQ+ +KE+ N
Sbjct: 322 VKEIQSQDNPLWIQYTKESQSKDQLPISNLGVKEIQSHDN 441
Score = 26.6 bits (57), Expect = 4.1
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 2/92 (2%)
Frame = +1
Query: 68 RYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGVKELQNLE 127
+Y K + ++QL P S L + ++ +QY ++ + +S LGVKE+Q+ +
Sbjct: 454 QYTKESQSNDQL--PISNLGAKEIQSQDNPLWIQYTKKSQSKDQLP-ISNLGVKEIQSQD 624
Query: 128 NQLEMSLKGVRMKKDQILTDE--IKELHQKGN 157
N L + DQ+ +KE+ N
Sbjct: 625 NPLWIQYTKESQSNDQLPISNLGVKEIQSHDN 720
>TC19118 weakly similar to UP|Q84YE8 (Q84YE8) Expressed protein-like
protein, partial (11%)
Length = 932
Score = 29.3 bits (64), Expect = 0.63
Identities = 35/133 (26%), Positives = 61/133 (45%), Gaps = 2/133 (1%)
Frame = +3
Query: 91 TEAASLRQQLQYLQE--CHRQLMGEGLSGLGVKELQNLENQLEMSLKGVRMKKDQILTDE 148
T+AASL ++L+ ++ C Q GL +E + L + E KGVR ++D ++ +
Sbjct: 396 TKAASLARELKSIKSDLCFMQER----CGLLEEENRRLRDGFE---KGVRPEEDDLVRLQ 554
Query: 149 IKELHQKGNLIHQENAELYKKMELIQKENAELQKKVYEARSTNEKDAASSPSCTIRNGYD 208
++ L + + + ENA L +EN L + V + ++ D + S I
Sbjct: 555 LEALLAEKSRLANENANLI-------RENQCLHQLVEYHQLASQDDLSESYEDVI----- 698
Query: 209 LQAISLQLSQPQP 221
Q + L S P P
Sbjct: 699 -QGMHLDFSSPPP 734
>TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%)
Length = 1742
Score = 29.3 bits (64), Expect = 0.63
Identities = 15/46 (32%), Positives = 24/46 (51%)
Frame = +3
Query: 183 KVYEARSTNEKDAASSPSCTIRNGYDLQAISLQLSQPQPQFSEPPA 228
KV E D ++ + +G D+ +ISL LS+P P F++ A
Sbjct: 798 KVCFGEDCPESDILAALDAAVEDGVDVISISLGLSEPPPFFNDSTA 935
>CN825477
Length = 466
Score = 27.7 bits (60), Expect = 1.8
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 5/48 (10%)
Frame = +1
Query: 185 YEARSTNEKDAASSPSCTIRNGYDL-----QAISLQLSQPQPQFSEPP 227
Y A T KDA P + GY Q Q + P PQ+S+PP
Sbjct: 67 YPAPDTYGKDAYPPPGYPPQQGYPPAGYPPQGYPPQYAPPPPQYSQPP 210
>TC17153 UP|AAQ87023 (AAQ87023) VDAC3.1, complete
Length = 1175
Score = 26.6 bits (57), Expect = 4.1
Identities = 12/39 (30%), Positives = 19/39 (47%)
Frame = +2
Query: 132 MSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKM 170
+ L +KKDQI +I L++ GNL + Y +
Sbjct: 260 LGLIATGLKKDQIFIGDINTLYKSGNLTVDVKVDTYSNV 376
>BP057683
Length = 469
Score = 26.2 bits (56), Expect = 5.4
Identities = 10/22 (45%), Positives = 16/22 (72%)
Frame = -3
Query: 18 QVTFSKRRNGLLKKAKELSILC 39
++ FSK +N + KK K +S+LC
Sbjct: 236 RIGFSKVQNSVRKKKKSISVLC 171
>TC9156 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fragment),
partial (3%)
Length = 850
Score = 26.2 bits (56), Expect = 5.4
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Frame = -2
Query: 47 VFSSTGKLYE---YASTSMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYL 103
+F ++ +LY YAS +++ VI YNK KE + P Q AAS Q+L
Sbjct: 762 LFLTSNQLYRHEIYASENLEGVI*TYNKRKE-----LFPG*RRALTQALAASTINNKQHL 598
Query: 104 QECH 107
H
Sbjct: 597 LPSH 586
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.129 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,192,008
Number of Sequences: 28460
Number of extensions: 34939
Number of successful extensions: 203
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 196
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 200
length of query: 241
length of database: 4,897,600
effective HSP length: 88
effective length of query: 153
effective length of database: 2,393,120
effective search space: 366147360
effective search space used: 366147360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0055.1


