

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0052b.4
(243 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
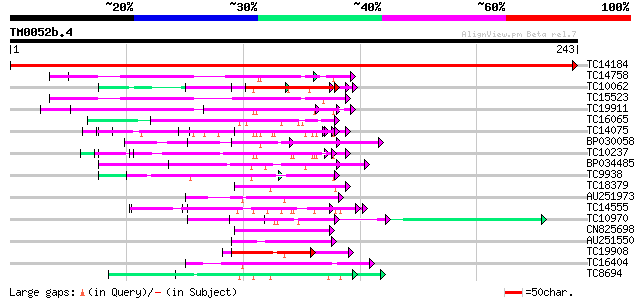
Score E
Sequences producing significant alignments: (bits) Value
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 524 e-150
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 67 2e-12
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 67 3e-12
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 65 1e-11
TC19911 weakly similar to PIR|A44805|A44805 eggshell protein pre... 61 2e-10
TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 61 2e-10
TC14075 homologue to UP|PRP2_MEDTR (Q40375) Repetitive proline-r... 59 6e-10
BP030058 54 2e-08
TC10237 similar to UP|NO75_SOYBN (P08297) Early nodulin 75 precu... 50 3e-07
BP034485 50 5e-07
TC9938 weakly similar to GB|AAM97131.1|22531255|AY136466 ribonuc... 49 8e-07
TC18379 similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich glycop... 48 1e-06
AU251973 48 2e-06
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 47 2e-06
TC10970 similar to GB|AAA32722.1|166570|ATH18RAB glycine rich pr... 47 2e-06
CN825698 47 3e-06
AU251550 47 3e-06
TC19908 similar to GB|CAA60022.1|791150|VURNEXT26 extensin-like ... 45 1e-05
TC16404 similar to UP|P93320 (P93320) Cdc2MsC protein, partial (... 45 1e-05
TC8694 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation ... 45 1e-05
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 524 bits (1349), Expect = e-150
Identities = 243/243 (100%), Positives = 243/243 (100%)
Frame = +3
Query: 1 MAEENRDRGLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYET 60
MAEENRDRGLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYET
Sbjct: 81 MAEENRDRGLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYET 260
Query: 61 GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG 120
GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG
Sbjct: 261 GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG 440
Query: 121 GGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYAL 180
GGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYAL
Sbjct: 441 GGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYAL 620
Query: 181 HEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
HEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF
Sbjct: 621 HEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 800
Query: 241 GRD 243
GRD
Sbjct: 801 GRD 809
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 67.4 bits (163), Expect = 2e-12
Identities = 50/136 (36%), Positives = 61/136 (44%), Gaps = 5/136 (3%)
Frame = +1
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DR T G+ +++++E +S+ E G G N T + + GG
Sbjct: 184 DRETGRSRGFGFVTFTSEEAM---------RSAIEGMNGNELDGRNITVNEAQARGGGGG 336
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGAG--GGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GG GGGYG GG GGY GGG GGGGG Y GGGGGGY
Sbjct: 337 GG-------------GRGGGGYGGGGGRREGGYGGGGGSRGYGGGGGGYG-GGGGGGY-- 468
Query: 136 GG---GGYGNDDSGNR 148
GG GGYG D G+R
Sbjct: 469 GGRREGGYGGDGGGSR 516
Score = 57.4 bits (137), Expect = 2e-09
Identities = 43/109 (39%), Positives = 44/109 (39%), Gaps = 2/109 (1%)
Frame = +1
Query: 26 GYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYN 85
G N T GG G Y GG E GY GGGG GY
Sbjct: 286 GRNITVNEAQARGGGGGGGGRGGGGYGGGGGRREGGY-------------GGGGGSRGYG 426
Query: 86 KTAYSTDEPSGGGYGTGGAGG--GYSTGGGYGTGGGGGGEYSTGGGGGG 132
GGGYG GG GG G GGYG G GGG Y +GGGGGG
Sbjct: 427 G--------GGGGYGGGGGGGYGG-RREGGYG-GDGGGSRYGSGGGGGG 543
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 67.0 bits (162), Expect = 3e-12
Identities = 36/67 (53%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGG-AGGGYSTGGGYGTGG--GGGGEYSTGGGGGG 132
GGG G Y GGGYG GG GGGY GGGYG G GGGG Y GG GGG
Sbjct: 279 GGGAGGGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGGGGYGGSGGYGGGGGYGGGGRGGG 458
Query: 133 YSTGGGG 139
GGGG
Sbjct: 459 RGGGGGG 479
Score = 64.3 bits (155), Expect = 2e-11
Identities = 34/57 (59%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Frame = +3
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTG---GGGGGYSTGGGGYGNDDSGNRR 149
GGG G GG GGG GGGYG GG GGG Y G GG GGY GGGGYG G R
Sbjct: 294 GGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGGGGYGGSGGYG-GGGGYGGGGRGGGR 461
Score = 63.9 bits (154), Expect = 2e-11
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGGY GGGYG GG GGG GGGYG GGG GG GGGGG Y
Sbjct: 306 GGGGY--------------GGGGYGGGGYGGGGRGGGGYGGGGGYGGSGGYGGGGG-YGG 440
Query: 136 GGGGYGNDDSG 146
GG G G G
Sbjct: 441 GGRGGGRGGGG 473
Score = 60.5 bits (145), Expect = 3e-10
Identities = 28/40 (70%), Positives = 28/40 (70%)
Frame = +3
Query: 102 GGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
GGAGGG GGGYG GG GGG Y GG GGG GGGGYG
Sbjct: 282 GGAGGGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGGGGYG 401
Score = 41.6 bits (96), Expect = 1e-04
Identities = 32/82 (39%), Positives = 32/82 (39%)
Frame = +3
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG GY Y GGGY G GGGGY G Y GGG
Sbjct: 297 GGRGGGGYGGGGY--GGGGYGGG------------GRGGGGYGGG---GGYGGSGGYGGG 425
Query: 99 YGTGGAGGGYSTGGGYGTGGGG 120
G GG G GGG G GGGG
Sbjct: 426 GGYGGGG----RGGGRGGGGGG 479
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 64.7 bits (156), Expect = 1e-11
Identities = 45/133 (33%), Positives = 55/133 (40%), Gaps = 4/133 (3%)
Frame = +2
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DR T G+ ++++++ K + E G G N T + + GG
Sbjct: 458 DRETGRSRGFGFVTFASEQSM---------KDAIEGMNGQDIDGRNVTVNEAQARGRGGG 610
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGY---- 133
GG GGGYG G GGY GGG GGGGG Y G GGGGY
Sbjct: 611 GG----------------GGGYGGGRREGGYGGGGGSRYSGGGGGGYG-GSGGGGYGGRR 739
Query: 134 STGGGGYGNDDSG 146
GGGGYG G
Sbjct: 740 EGGGGGYGGSREG 778
>TC19911 weakly similar to PIR|A44805|A44805 eggshell protein precursor -
fluke (Schistosoma haematobium) {Schistosoma
haematobium;} , partial (24%)
Length = 350
Score = 61.2 bits (147), Expect = 2e-10
Identities = 40/120 (33%), Positives = 49/120 (40%)
Frame = +1
Query: 14 HKNEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEP 73
H N+ R +D E G ++ + SGG YD + +GGGY G + Y
Sbjct: 55 HGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGG--RTGGGYGDGGHSAGGYGDHGG 228
Query: 74 NSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGY 133
SGGG GGGYG GG G GGYG GG G Y G GG+
Sbjct: 229 RSGGG-----------------GGGYGDGGLSSG---SGGYGDGGRSGVGYGDGXLSGGH 348
Score = 60.1 bits (144), Expect = 3e-10
Identities = 39/117 (33%), Positives = 53/117 (44%), Gaps = 1/117 (0%)
Frame = +1
Query: 27 YNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNK 86
+ ++ +T GG + + ++ S E G ET + S D+ N GG
Sbjct: 10 FENSAGNTGHSGGGSHGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGGR------- 168
Query: 87 TAYSTDEPSGGGYGTGG-AGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGN 142
+GGGYG GG + GGY GG GGGGG G GG S+G GGYG+
Sbjct: 169 --------TGGGYGDGGHSAGGYGDHGGRSGGGGGGY------GDGGLSSGSGGYGD 297
Score = 39.3 bits (90), Expect = 6e-04
Identities = 28/76 (36%), Positives = 35/76 (45%), Gaps = 11/76 (14%)
Frame = +1
Query: 84 YNKTAYSTDEPSGGGYGTGGA------GGGYSTGGGYGTGGGGGGEYSTGGG--GGGYST 135
+ +A +T GG +G GG S G+G GGG + + GG GGGY
Sbjct: 10 FENSAGNTGHSGGGSHGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGGRTGGGYGD 189
Query: 136 GG---GGYGNDDSGNR 148
GG GGYG D G R
Sbjct: 190 GGHSAGGYG--DHGGR 231
>TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (20%)
Length = 608
Score = 61.2 bits (147), Expect = 2e-10
Identities = 50/133 (37%), Positives = 52/133 (38%), Gaps = 25/133 (18%)
Frame = -1
Query: 34 TDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDE 93
TD PSGG+ G S E S + P GG G
Sbjct: 467 TDGPSGGEGSEGQHSHSTERSA--------PPPQQAAAAPGGGGPG-------------- 354
Query: 94 PSGG-GYG---TGGAGGGYSTGGGYGTGGGGGG-------------EY--------STGG 128
P GG GYG GG GGGY GGG G GGGG G EY S G
Sbjct: 353 PQGGRGYGGPPQGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYGGPPEYQGRGRGGPSQQG 174
Query: 129 GGGGYSTGGGGYG 141
G GGYS GGGGYG
Sbjct: 173 GRGGYSGGGGGYG 135
Score = 46.2 bits (108), Expect = 5e-06
Identities = 32/83 (38%), Positives = 35/83 (41%), Gaps = 2/83 (2%)
Frame = -1
Query: 61 GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYG--TGG 118
G+ TD P+ G G + T S P GG G G G GYG G
Sbjct: 494 GFIMVRKRRTDGPSGGEGSEGQHSHSTERSAPPPQQAAAAPGGGGPGPQGGRGYGGPPQG 315
Query: 119 GGGGEYSTGGGGGGYSTGGGGYG 141
G GG Y GGGGG GGGG G
Sbjct: 314 GRGGGY--GGGGGP-GYGGGGRG 255
>TC14075 homologue to UP|PRP2_MEDTR (Q40375) Repetitive proline-rich cell
wall protein 2 precursor, partial (98%)
Length = 1524
Score = 59.3 bits (142), Expect = 6e-10
Identities = 50/135 (37%), Positives = 65/135 (48%), Gaps = 26/135 (19%)
Frame = -2
Query: 32 YSTDEPSGGDYDSGYKKSSYETSG--------GGYETGYNKTSSYSTDEPNSGG------ 77
Y+ +GG Y G+ T G GG+ TG T +ST +GG
Sbjct: 533 YTGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL 354
Query: 78 --GGYDSGYNKTA-YSTDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG- 128
GG+ +G T +ST GG+ TGG GG+STGG G+ TGG G +STGG
Sbjct: 353 *TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGL 174
Query: 129 GGGGYSTGG---GGY 140
GG+STGG GG+
Sbjct: 173 YTGGFSTGGL*TGGF 129
Score = 58.9 bits (141), Expect = 8e-10
Identities = 48/121 (39%), Positives = 62/121 (50%), Gaps = 18/121 (14%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG--------GGYDSGYNKTA- 88
+GG Y G+ T GG+ TG T +ST +GG GG+ +G T
Sbjct: 545 TGGLYTGGFSTGGLYT--GGFSTGGLYTGGFSTGGL*TGGFSTGGL*TGGFSTGGLYTGG 372
Query: 89 YSTDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GG 139
+ST GG+ TGG GG+STGG G+ TGG G +STGG GG+STGG GG
Sbjct: 371 FSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGG 192
Query: 140 Y 140
+
Sbjct: 191 F 189
Score = 58.2 bits (139), Expect = 1e-09
Identities = 51/138 (36%), Positives = 65/138 (46%), Gaps = 23/138 (16%)
Frame = -2
Query: 32 YSTDEPSGGDYDSGYKKSSYETSG--------GGYETGYNKTSSYSTDEPNSGG------ 77
Y+ +GG Y G+ T G GG+ TG T +ST +GG
Sbjct: 503 YTGGFSTGGLYTGGFSTGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGL 324
Query: 78 --GGYDSGYNKTA-YSTDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG- 128
GG+ +G T +ST GG+ TGG GG+STGG G+ TGG G +STGG
Sbjct: 323 YTGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGL 144
Query: 129 GGGGYSTGGGGYGNDDSG 146
GG+STGG G SG
Sbjct: 143 *TGGFSTGGL*IGGVYSG 90
Score = 57.8 bits (138), Expect = 2e-09
Identities = 49/135 (36%), Positives = 64/135 (47%), Gaps = 26/135 (19%)
Frame = -2
Query: 32 YSTDEPSGGDYDSGYKKSSYETSG--------GGYETGYNKTSSYSTDEPNSGG------ 77
Y+ +GG Y G+ T G GG+ G T +ST +GG
Sbjct: 743 YTGGFSTGGLYTGGFSTGGLYTGGL*TGGL*TGGFSAGGL*TGGFSTGGLYTGGFSTGGL 564
Query: 78 --GGYDSGYNKTA-YSTDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG- 128
GG+ +G T +ST GG+ TGG GG+STGG G+ TGG G +STGG
Sbjct: 563 *TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TGGFSTGGL*TGGFSTGGL 384
Query: 129 GGGGYSTGG---GGY 140
GG+STGG GG+
Sbjct: 383 YTGGFSTGGL*TGGF 339
Score = 57.8 bits (138), Expect = 2e-09
Identities = 48/124 (38%), Positives = 62/124 (49%), Gaps = 21/124 (16%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG--------GGYDSGYNK 86
+GG G + T G GG+ TG T +ST +GG GG+ +G
Sbjct: 800 TGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGL*TGGL*TGGFSAGGL* 621
Query: 87 TA-YSTDEPSGGGYGTGGA-GGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG-- 137
T +ST GG+ TGG GG+STG GG+ TGG G +STGG GG+STGG
Sbjct: 620 TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGL* 441
Query: 138 -GGY 140
GG+
Sbjct: 440 TGGF 429
Score = 56.2 bits (134), Expect = 5e-09
Identities = 48/119 (40%), Positives = 61/119 (50%), Gaps = 16/119 (13%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTA-YS 90
+GG Y G+ T G GG TG T T ++GG GG+ +G T +S
Sbjct: 755 TGGLYTGGFSTGGLYTGGFSTGGLYTGGL*TGGL*TGGFSAGGL*TGGFSTGGLYTGGFS 576
Query: 91 TDEPSGGGYGTGGA-GGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
T GG+ TGG GG+STG GG+ TGG G +STGG GG+STGG GG+
Sbjct: 575 TGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TGGFSTGGL*TGGF 399
Score = 54.7 bits (130), Expect = 1e-08
Identities = 48/125 (38%), Positives = 59/125 (46%), Gaps = 22/125 (17%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG--------GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAY 89
+GG Y G+ T G GG+ TG T +ST GG Y G++
Sbjct: 845 TGGLYTGGFSTGGL*TGGFSTGGL*TGGFSTGGLYTGGFST------GGLYTGGFSTGGL 684
Query: 90 STDEPSGGGYGTGG--AGG----GYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG- 137
T GG TGG AGG G+STG GG+ TGG G +STGG GG+STGG
Sbjct: 683 YTGGL*TGGL*TGGFSAGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGL 504
Query: 138 --GGY 140
GG+
Sbjct: 503 YTGGF 489
Score = 52.8 bits (125), Expect = 5e-08
Identities = 44/109 (40%), Positives = 55/109 (50%), Gaps = 10/109 (9%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYSTDEP 94
+GG Y G+ T GG+ TG T +ST +GG GG +G +ST
Sbjct: 395 TGGLYTGGFSTGGL*T--GGFSTGGLYTGGFSTGGL*TGGFSTGGLYTG----GFSTGGL 234
Query: 95 SGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG--GGGGYSTG 136
GG+ TGG GG+STGG G+ TGG G +STGG GG YS G
Sbjct: 233 YTGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TGGFSTGGL*IGGVYSGG 87
Score = 52.8 bits (125), Expect = 5e-08
Identities = 48/123 (39%), Positives = 60/123 (48%), Gaps = 21/123 (17%)
Frame = -2
Query: 39 GGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTA---- 88
GG G + T G GG+ TG T +ST +GG GG +G T
Sbjct: 1067 GGFSTGGL*TGGFSTGGL*TGGFSTGGL*TGGFSTGGLYTGGL*TGGLYTGGL*TGGL*T 888
Query: 89 --YSTDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG--- 137
+ST GG+ TGG GG+STGG G+ TGG G +STGG GG+STGG
Sbjct: 887 GGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGLYT 708
Query: 138 GGY 140
GG+
Sbjct: 707 GGF 699
Score = 50.4 bits (119), Expect = 3e-07
Identities = 47/130 (36%), Positives = 58/130 (44%), Gaps = 27/130 (20%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG--------GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAY 89
+GG Y G T G GG+ TG T +ST GG Y G++
Sbjct: 965 TGGLYTGGL*TGGLYTGGL*TGGL*TGGFSTGGL*TGGFST------GGLYTGGFSTGGL 804
Query: 90 STDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG---GG--------GGY 133
T GG+ TGG GG+STGG G+ TGG G +STGG GG GG+
Sbjct: 803 *T-----GGFSTGGL*TGGFSTGGLYTGGFSTGGLYTGGFSTGGLYTGGL*TGGL*TGGF 639
Query: 134 STGG---GGY 140
S GG GG+
Sbjct: 638 SAGGL*TGGF 609
Score = 48.5 bits (114), Expect = 1e-06
Identities = 45/125 (36%), Positives = 56/125 (44%), Gaps = 22/125 (17%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG--------GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAY 89
+GG Y G+ T G GG+ TG T +ST GG G++
Sbjct: 1145 TGGLYTGGFSTGGL*TGGFSTGGL*IGGFSTGGL*TGGFST------GGL*TGGFSTGGL 984
Query: 90 STDEPSGGGYGTGG--AGGGYSTG--------GGYGTGGGGGGEYSTGG-GGGGYSTGG- 137
T S GG TGG GG Y+ G GG+ TGG G +STGG GG+STGG
Sbjct: 983 *TGGFSTGGLYTGGL*TGGLYTGGL*TGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL 804
Query: 138 --GGY 140
GG+
Sbjct: 803 *TGGF 789
Score = 46.6 bits (109), Expect = 4e-06
Identities = 47/122 (38%), Positives = 59/122 (47%), Gaps = 26/122 (21%)
Frame = -2
Query: 45 GYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTA-YSTDEPSGG 97
G+ + T G GG+ TG T +ST GG GG+ +G T +ST G
Sbjct: 1169 GFPYGGFSTGGLYTGGFSTGGL*TGGFST-----GGL*IGGFSTGGL*TGGFSTGGL*TG 1005
Query: 98 GYGTGGA-GGGYSTGGGYGTGGGGGGEYS---------TGG---GG---GGYSTGG---G 138
G+ TGG GG+STGG Y G GG Y+ TGG GG GG+STGG G
Sbjct: 1004 GFSTGGL*TGGFSTGGLYTGGL*TGGLYTGGL*TGGL*TGGFSTGGL*TGGFSTGGLYTG 825
Query: 139 GY 140
G+
Sbjct: 824 GF 819
Score = 46.2 bits (108), Expect = 5e-06
Identities = 34/77 (44%), Positives = 42/77 (54%), Gaps = 14/77 (18%)
Frame = -2
Query: 78 GGYDS-----GYNKTAYSTDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTG 127
GGY G+ +ST GG+ TGG GG+STGG G+ TGG G +STG
Sbjct: 1199 GGYPG*PPYGGFPYGGFSTGGLYTGGFSTGGL*TGGFSTGGL*IGGFSTGGL*TGGFSTG 1020
Query: 128 G-GGGGYSTGG---GGY 140
G GG+STGG GG+
Sbjct: 1019 GL*TGGFSTGGL*TGGF 969
Score = 45.4 bits (106), Expect = 9e-06
Identities = 35/75 (46%), Positives = 43/75 (56%), Gaps = 10/75 (13%)
Frame = -2
Query: 73 PNSGG---GGYDSGYNKTA-YSTDEPSGGGYGTGGA-GGGYSTG----GGYGTGGGGGGE 123
P GG GG+ +G T +ST GG+ TGG GG+STG GG+ TGG G
Sbjct: 1181 PPYGGFPYGGFSTGGLYTGGFSTGGL*TGGFSTGGL*IGGFSTGGL*TGGFSTGGL*TGG 1002
Query: 124 YSTGG-GGGGYSTGG 137
+STGG GG+STGG
Sbjct: 1001 FSTGGL*TGGFSTGG 957
Score = 38.9 bits (89), Expect = 8e-04
Identities = 28/56 (50%), Positives = 32/56 (57%), Gaps = 11/56 (19%)
Frame = -2
Query: 97 GGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG---GG---GGYSTGGGGYG 141
GG+ TGG GG+STGG G+ TGG G +STGG GG GG TGG G
Sbjct: 1157 GGFSTGGLYTGGFSTGGL*TGGFSTGGL*IGGFSTGGL*TGGFSTGGL*TGGFSTG 990
Score = 37.0 bits (84), Expect = 0.003
Identities = 28/59 (47%), Positives = 34/59 (57%), Gaps = 12/59 (20%)
Frame = -2
Query: 94 PSGGGYG----TGGAGGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
P+GG G G GG+STG GG+ TGG G +STGG GG+STGG GG+
Sbjct: 1205 PAGGYPG*PPYGGFPYGGFSTGGLYTGGFSTGGL*TGGFSTGGL*IGGFSTGGL*TGGF 1029
>BP030058
Length = 455
Score = 54.3 bits (129), Expect = 2e-08
Identities = 31/66 (46%), Positives = 34/66 (50%)
Frame = +1
Query: 77 GGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG 136
GGG D G G +G GG GGG GG G GGGGGG+ GGGGG G
Sbjct: 247 GGGGDKGLQFCGQE------GYFGGGGEGGG----GGGGDGGGGGGDGHGSGGGGGEGDG 396
Query: 137 GGGYGN 142
GGG G+
Sbjct: 397 GGGGGS 414
Score = 48.1 bits (113), Expect = 1e-06
Identities = 29/66 (43%), Positives = 34/66 (50%), Gaps = 1/66 (1%)
Frame = +1
Query: 96 GGGYGTGGAGG-GYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDY 154
GG G G GY GGG G GGGGGG+ GGG G S GGGG G+ G +V
Sbjct: 253 GGDKGLQFCGQEGYFGGGGEG-GGGGGGDGGGGGGDGHGSGGGGGEGDGGGGGGSPQVSP 429
Query: 155 KEEEKH 160
+ + H
Sbjct: 430 QGHDPH 447
Score = 39.7 bits (91), Expect = 5e-04
Identities = 28/73 (38%), Positives = 31/73 (42%)
Frame = +1
Query: 50 SYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYS 109
+Y GGG + G + E GGGG G GGG G G GG
Sbjct: 232 TYMILGGGGDKGLQ----FCGQEGYFGGGGEGGG------GGGGDGGGGGGDGHGSGG-- 375
Query: 110 TGGGYGTGGGGGG 122
GGG G GGGGGG
Sbjct: 376 -GGGEGDGGGGGG 411
Score = 36.6 bits (83), Expect = 0.004
Identities = 26/61 (42%), Positives = 26/61 (42%), Gaps = 10/61 (16%)
Frame = +1
Query: 103 GAGGGYSTGGGYGTGG----------GGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEV 152
G G Y GG G G GGGGE GGGGGG GGGG SG E
Sbjct: 220 GCPGTYMILGGGGDKGLQFCGQEGYFGGGGE---GGGGGGGDGGGGGGDGHGSGGGGGEG 390
Query: 153 D 153
D
Sbjct: 391 D 393
Score = 28.1 bits (61), Expect = 1.4
Identities = 18/48 (37%), Positives = 20/48 (41%), Gaps = 7/48 (14%)
Frame = +1
Query: 107 GYSTGGGYGTGGGGG-------GEYSTGGGGGGYSTGGGGYGNDDSGN 147
G G Y GGGG G+ GGGG GGGG G G+
Sbjct: 214 GLGCPGTYMILGGGGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGGD 357
>TC10237 similar to UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75)
(NGM-75), partial (49%)
Length = 803
Score = 50.4 bits (119), Expect = 3e-07
Identities = 39/92 (42%), Positives = 47/92 (50%), Gaps = 5/92 (5%)
Frame = -1
Query: 54 SGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGA--GGGYSTG 111
SGGG+ G Y+ +GG G +Y E +GGG+ GG+ GGG+S G
Sbjct: 383 SGGGFS*G----G*YTGGR**NGG*GVGCPVGGLSYG-GEYTGGGFS*GGSYSGGGFSYG 219
Query: 112 GGYGTGGGGGGEYSTGGG---GGGYSTGGGGY 140
GGY GG G TGGG GG YS GGG Y
Sbjct: 218 GGYAGGGFQYGGRYTGGGFS*GGSYS-GGGWY 126
Score = 45.1 bits (105), Expect = 1e-05
Identities = 43/114 (37%), Positives = 51/114 (44%), Gaps = 4/114 (3%)
Frame = -1
Query: 37 PSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG 96
P GG G+ + GG+ G + +S SGGG G Y+
Sbjct: 491 PGGGS*SGGFS*GG--S*MGGFSWGGS*MGGFS*GGWYSGGGFS*GG*----YTGGR**N 330
Query: 97 GGYGTGGAGGGYSTGGGYGTGGGG--GGEYSTGGG--GGGYSTGGGGYGNDDSG 146
GG G G GG S GG Y TGGG GG YS GG GGGY+ GG YG +G
Sbjct: 329 GG*GVGCPVGGLSYGGEY-TGGGFS*GGSYSGGGFSYGGGYAGGGFQYGGRYTG 171
Score = 45.1 bits (105), Expect = 1e-05
Identities = 40/96 (41%), Positives = 46/96 (47%), Gaps = 7/96 (7%)
Frame = -1
Query: 52 ETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGA--GGGYS 109
E +GGG+ G SYS GG Y GY + GG Y GG GG YS
Sbjct: 281 EYTGGGFS*G----GSYS-----GGGFSYGGGYAGGGFQY----GGRYTGGGFS*GGSYS 141
Query: 110 TGGGYGTGGG-GGGEYSTGG-GGGGYSTGG---GGY 140
GG Y GG GG YS GG GG+S+GG GG+
Sbjct: 140 GGGWYCDGGFL*GGWYSGGGFSWGGFSSGGFSKGGF 33
Score = 43.9 bits (102), Expect = 3e-05
Identities = 36/102 (35%), Positives = 44/102 (42%), Gaps = 3/102 (2%)
Frame = -1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG+Y G SGGG+ G Y+ GG Y Y +S GG
Sbjct: 287 GGEYTGGGFS*GGSYSGGGFSYG----GGYA-----GGGFQYGGRYTGGGFS*----GGS 147
Query: 99 YGTGG--AGGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGG 137
Y GG GG+ GG Y GG G +S+GG GG+S GG
Sbjct: 146 YSGGGWYCDGGFL*GGWYSGGGFSWGGFSSGGFSKGGFS*GG 21
Score = 40.8 bits (94), Expect = 2e-04
Identities = 35/109 (32%), Positives = 43/109 (39%), Gaps = 2/109 (1%)
Frame = -1
Query: 31 SYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYS 90
SY + GG G + GGGY G + T S GG Y
Sbjct: 293 SYGGEYTGGGFS*GGSYSGGGFSYGGGYAGGGFQYGGRYTGGGFS*GGSYS--------- 141
Query: 91 TDEPSGGGYGTGG-AGGGYSTGGGYGTGGGGGGEYSTGGGG-GGYSTGG 137
GG Y GG GG+ +GGG+ GG G +S GG GG+ GG
Sbjct: 140 ----GGGWYCDGGFL*GGWYSGGGFSWGGFSSGGFSKGGFS*GGWYLGG 6
Score = 37.7 bits (86), Expect = 0.002
Identities = 37/102 (36%), Positives = 43/102 (41%), Gaps = 31/102 (30%)
Frame = -1
Query: 76 GGGGYDSGYN-KTAYSTDEPSGGGYGTGGA-GGGYSTGGGYGTGG---GG---------- 120
GGG Y G + +S GG+ GG+ GG+S GG Y GG GG
Sbjct: 506 GGGLYPGGGS*SGGFS*GGS*MGGFSWGGS*MGGFS*GGWYSGGGFS*GG*YTGGR**NG 327
Query: 121 -------------GGEYSTGGG---GGGYSTGGGGYGNDDSG 146
GGEY TGGG GG YS GG YG +G
Sbjct: 326 G*GVGCPVGGLSYGGEY-TGGGFS*GGSYSGGGFSYGGGYAG 204
Score = 35.4 bits (80), Expect = 0.009
Identities = 24/52 (46%), Positives = 28/52 (53%), Gaps = 8/52 (15%)
Frame = -1
Query: 94 PSGGGYGTGGAG--GGYSTGG----GYGTGGGGGGEYSTGG--GGGGYSTGG 137
P GGG GG GG+S GG G+ GG G +S GG GGG+S GG
Sbjct: 512 PYGGGLYPGGGS*SGGFS*GGS*MGGFSWGGS*MGGFS*GGWYSGGGFS*GG 357
>BP034485
Length = 485
Score = 49.7 bits (117), Expect = 5e-07
Identities = 39/124 (31%), Positives = 53/124 (42%), Gaps = 20/124 (16%)
Frame = +3
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GGD S KK + + +GGG+ + S + GGGG + NK + ++ +GGG
Sbjct: 48 GGDSSSDKKKKTKDKNGGGFLVKFLGLGKKSK-KGGGGGGGETANKNK---NKNKNNGGG 215
Query: 99 YGTG------GAGGGY--------------STGGGYGTGGGGGGEYSTGGGGGGYSTGGG 138
G G GGG S G G GGGGGG+ + G G+ GG
Sbjct: 216 NNKGKEGKKGGGGGGKLDKIDFDFQDFDIPSHGKSGGKGGGGGGKGNKNGHDHGHGIGGN 395
Query: 139 GYGN 142
GN
Sbjct: 396 MNGN 407
Score = 42.7 bits (99), Expect = 6e-05
Identities = 30/92 (32%), Positives = 43/92 (46%), Gaps = 6/92 (6%)
Frame = +3
Query: 69 STDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYS--- 125
S + GGGG DS +K + D+ +GGG+ G G + G GGGGGGE +
Sbjct: 18 SKEGKQKGGGGGDSSSDKKKKTKDK-NGGGFLVKFLGLGKKSKKG---GGGGGGETANKN 185
Query: 126 ---TGGGGGGYSTGGGGYGNDDSGNRRDEVDY 154
GGG + G G G + D++D+
Sbjct: 186 KNKNKNNGGGNNKGKEGKKGGGGGGKLDKIDF 281
>TC9938 weakly similar to GB|AAM97131.1|22531255|AY136466
ribonucleoprotein-like {Arabidopsis thaliana;}, partial
(9%)
Length = 633
Score = 48.9 bits (115), Expect = 8e-07
Identities = 38/102 (37%), Positives = 44/102 (42%), Gaps = 11/102 (10%)
Frame = +1
Query: 51 YETSGGGYETGYNKTSSYSTDEPNSG-----GGGYDSGYNKTAYSTDEPSGGGYGTG-GA 104
Y + GGY GY +P+ G GG + GY+ Y S G YG G A
Sbjct: 43 YGSEFGGYG-GYAGAMGPYRGDPSLGFAGRYGGSFSRGYDLGGYGGASESYGAYGAGAAA 219
Query: 105 GGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGG-----GGYG 141
GGG S G Y GG + S GG GG S G GGYG
Sbjct: 220 GGGSSAAGAY----QGGYDASLAGGYGGASGGSFYGSRGGYG 333
Score = 40.8 bits (94), Expect = 2e-04
Identities = 25/79 (31%), Positives = 31/79 (38%)
Frame = +1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG + GY Y GG E+ + + +S G Y GY D GG
Sbjct: 133 GGSFSRGYDLGGY---GGASESYGAYGAGAAAGGGSSAAGAYQGGY-------DASLAGG 282
Query: 99 YGTGGAGGGYSTGGGYGTG 117
YG G Y + GGYG G
Sbjct: 283 YGGASGGSFYGSRGGYGAG 339
Score = 33.9 bits (76), Expect = 0.026
Identities = 24/70 (34%), Positives = 30/70 (42%), Gaps = 8/70 (11%)
Frame = +1
Query: 85 NKTAYSTDEPSGGGYGT--GGAGGGYSTGGGY------GTGGGGGGEYSTGGGGGGYSTG 136
+ AY+ + GGYG+ GG GG G Y G G GG +S G GGY
Sbjct: 1 SSAAYAGRSGAYGGYGSEFGGYGGYAGAMGPYRGDPSLGFAGRYGGSFSRGYDLGGYGGA 180
Query: 137 GGGYGNDDSG 146
YG +G
Sbjct: 181 SESYGAYGAG 210
>TC18379 similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich glycoprotein
DZ-HRGP precursor, partial (8%)
Length = 584
Score = 48.1 bits (113), Expect = 1e-06
Identities = 28/63 (44%), Positives = 30/63 (47%), Gaps = 13/63 (20%)
Frame = -1
Query: 97 GGYGTGGAGGGYST--------GGGYGTGGGGGGEYSTGGGGGGYSTGGG-----GYGND 143
GG G GG GG GGG GGG G GGGGGG + GGG GYGN+
Sbjct: 386 GGAGLGGRGGKVDVKRGGP*GRGGGAAQYGGGAGGAGAGGGGGGGAIGGG*NWC*GYGNE 207
Query: 144 DSG 146
G
Sbjct: 206 GYG 198
Score = 37.7 bits (86), Expect = 0.002
Identities = 29/71 (40%), Positives = 31/71 (42%), Gaps = 3/71 (4%)
Frame = -1
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGG---G 132
GG G K P G G G GGG GG G GGGGGG + GGG G
Sbjct: 386 GGAGLGGRGGKVDVKRGGP*GRGGGAAQYGGG---AGGAGAGGGGGG-GAIGGG*NWC*G 219
Query: 133 YSTGGGGYGND 143
Y G GYG +
Sbjct: 218 Y--GNEGYGEN 192
Score = 28.1 bits (61), Expect = 1.4
Identities = 15/34 (44%), Positives = 17/34 (49%)
Frame = -1
Query: 95 SGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGG 128
+GGG G G GGG + GYG G G GG
Sbjct: 275 AGGGGGGGAIGGG*NWC*GYGNEGYGENSIP*GG 174
>AU251973
Length = 205
Score = 47.8 bits (112), Expect = 2e-06
Identities = 34/76 (44%), Positives = 37/76 (47%), Gaps = 8/76 (10%)
Frame = -2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGG----YGTGGAGGGYSTGGGYGT----GGGGGGEYSTG 127
GGGG + Y GGG YG GG G Y GGG G GGGGG EY G
Sbjct: 201 GGGGDEYLYG---------GGGGDEYLYG-GGGGDEYLYGGGGGDEYL*GGGGGDEYLYG 52
Query: 128 GGGGGYSTGGGGYGND 143
GGGG GGG G++
Sbjct: 51 GGGGDEYL*GGGGGDE 4
Score = 36.2 bits (82), Expect = 0.005
Identities = 16/27 (59%), Positives = 18/27 (66%)
Frame = -2
Query: 117 GGGGGGEYSTGGGGGGYSTGGGGYGND 143
GGGGG EY GGGGG GGG G++
Sbjct: 204 GGGGGDEYLYGGGGGDEYLYGGGGGDE 124
Score = 34.7 bits (78), Expect = 0.015
Identities = 21/47 (44%), Positives = 23/47 (48%), Gaps = 8/47 (17%)
Frame = -2
Query: 105 GGGYSTGGGYGTGGG--------GGGEYSTGGGGGGYSTGGGGYGND 143
GGG YG GGG GG EY GGGGG GGG G++
Sbjct: 204 GGGGGDEYLYGGGGGDEYLYGGGGGDEYLYGGGGGDEYL*GGGGGDE 64
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 47.4 bits (111), Expect = 2e-06
Identities = 34/87 (39%), Positives = 37/87 (42%), Gaps = 13/87 (14%)
Frame = -1
Query: 77 GGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG-----GGEYSTGGG-- 129
G G +G +A GGG GG GG GGG G GG GG TGGG
Sbjct: 519 GAGEVAGTGVSAGGGVAAGGGGVAGGGEKGGGEAGGGVLAGAGGGVD*TGGGEDTGGGED 340
Query: 130 --GGGYSTGGG----GYGNDDSGNRRD 150
GGG GGG G G D +G D
Sbjct: 339 *AGGGVDAGGGED*TGGGED*AGGGED 259
Score = 44.7 bits (104), Expect = 1e-05
Identities = 36/96 (37%), Positives = 42/96 (43%), Gaps = 9/96 (9%)
Frame = -1
Query: 53 TSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGG----AGGGY 108
++GGG G + GGG +G A + +GGG TGG AGGG
Sbjct: 489 SAGGGVAAGGGGVAGGGEKGGGEAGGGVLAG----AGGGVD*TGGGEDTGGGED*AGGGV 322
Query: 109 STGGGYG-TGGG----GGGEYSTGGGGGGYSTGGGG 139
GGG TGGG GGGE GG G GG G
Sbjct: 321 DAGGGED*TGGGED*AGGGED*AAGG*GVVGVGGAG 214
Score = 43.1 bits (100), Expect = 4e-05
Identities = 35/105 (33%), Positives = 46/105 (43%), Gaps = 6/105 (5%)
Frame = -1
Query: 52 ETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTG 111
E G G G ++T+ + +G G D + E +G G AGGG + G
Sbjct: 624 ERLGPGASDGESETTGANA---GAGAGAGDLLFTLAGAGAGEVAGTGVS---AGGGVAAG 463
Query: 112 GGYGTGGG--GGGEYSTGGGGGGYSTGGGGY----GNDDSGNRRD 150
GG GGG GGGE GGG + GGG G +D+G D
Sbjct: 462 GGGVAGGGEKGGGE----AGGGVLAGAGGGVD*TGGGEDTGGGED 340
Score = 39.3 bits (90), Expect = 6e-04
Identities = 35/99 (35%), Positives = 41/99 (41%), Gaps = 20/99 (20%)
Frame = -1
Query: 75 SGGGGYDSGYNKTAYSTDEPSG--GGYGTGGAGGGYS-TGGGYGTGGG------------ 119
S GGG +G A ++ G GG GAGGG TGGG TGGG
Sbjct: 489 SAGGGVAAGGGGVAGGGEKGGGEAGGGVLAGAGGGVD*TGGGEDTGGGED*AGGGVDAGG 310
Query: 120 -----GGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVD 153
GGGE GGG + G G G +G E+D
Sbjct: 309 GED*TGGGED*AGGGED*AAGG*GVVGVGGAGEVVGELD 193
Score = 30.8 bits (68), Expect = 0.22
Identities = 26/85 (30%), Positives = 36/85 (41%)
Frame = -1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG G +K E +GGG G + ++GGG +G A ++ +GGG
Sbjct: 462 GGGVAGGGEKGGGE-AGGGVLAGAGGGVD*TGGGEDTGGGED*AGGGVDAGGGED*TGGG 286
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGE 123
G G + GG G GG GE
Sbjct: 285 ED*AGGGED*AAGG*GVVGVGGAGE 211
>TC10970 similar to GB|AAA32722.1|166570|ATH18RAB glycine rich protein
{Arabidopsis thaliana;} , partial (25%)
Length = 697
Score = 47.4 bits (111), Expect = 2e-06
Identities = 27/49 (55%), Positives = 29/49 (59%), Gaps = 2/49 (4%)
Frame = +2
Query: 95 SGGGYGTGGAGGGYSTGG--GYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
+G GYGTGG G Y T G G+GT G G G Y TGG G Y T G G G
Sbjct: 8 TGTGYGTGGHGTDYGTTGTHGHGTTGTGTG-YGTGGHGTEYGTTGTGGG 151
Score = 44.7 bits (104), Expect = 1e-05
Identities = 36/108 (33%), Positives = 44/108 (40%), Gaps = 21/108 (19%)
Frame = +2
Query: 77 GGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGG---YGTGGGGGG----------- 122
G GY +G + T Y T G +GT G G GY TGG YGT G GGG
Sbjct: 11 GTGYGTGGHGTDYGTTGTHG--HGTTGTGTGYGTGGHGTEYGTTGTGGGVMDKIKEKLPC 184
Query: 123 -------EYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKK 163
EY +GG G +T G +G E+HH+K
Sbjct: 185 AGSTATHEYGSGGTTGTTTTTTGHHG----------------EQHHEK 280
Score = 39.3 bits (90), Expect = 6e-04
Identities = 31/121 (25%), Positives = 42/121 (34%)
Frame = +2
Query: 110 TGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGE 169
TG GYGTGG G +TG G G + G GYG G E
Sbjct: 8 TGTGYGTGGHGTDYGTTGTHGHGTTGTGTGYGTGGHGT---------------------E 124
Query: 170 LGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEE 229
GTT G + + + + H+ G +HHEKK ++ +
Sbjct: 125 YGTTGTGGGVMDKIKEKLPCAGSTATHEYGSGGTTGTTTTTTGHHGEQHHEKKGIMDKIK 304
Query: 230 E 230
E
Sbjct: 305 E 307
Score = 38.5 bits (88), Expect = 0.001
Identities = 24/49 (48%), Positives = 26/49 (52%), Gaps = 5/49 (10%)
Frame = +2
Query: 103 GAGGGYSTGG---GYGTGGGGGGEYSTGGGGGGYSTGGGG--YGNDDSG 146
G G GY TGG YGT G G + T G G GY TGG G YG +G
Sbjct: 5 GTGTGYGTGGHGTDYGTTGTHG--HGTTGTGTGYGTGGHGTEYGTTGTG 145
Score = 32.7 bits (73), Expect = 0.058
Identities = 29/104 (27%), Positives = 38/104 (35%)
Frame = +2
Query: 21 TDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGY 80
T T TGY + TD + G + G + GG+ T Y T +GGG
Sbjct: 2 TGTGTGYGTGGHGTDYGTTGTHGHGTTGTGTGYGTGGHGTEYGTT--------GTGGGVM 157
Query: 81 DSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEY 124
D K P G T G G +TG T G G ++
Sbjct: 158 DKIKEKL------PCAGSTATHEYGSGGTTGTTTTTTGHHGEQH 271
>CN825698
Length = 704
Score = 47.0 bits (110), Expect = 3e-06
Identities = 23/43 (53%), Positives = 25/43 (57%)
Frame = -1
Query: 97 GGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGG 139
GG G + GG S GGG G GGGGG E G GGG S+ GG
Sbjct: 206 GGGGRSESEGGGSDGGGGGGGGGGGAEAEDGDGGGDLSSESGG 78
Score = 37.7 bits (86), Expect = 0.002
Identities = 20/39 (51%), Positives = 21/39 (53%)
Frame = -1
Query: 105 GGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGND 143
GGG S G G+ GGGGG GGGGGG G G D
Sbjct: 203 GGGRSESEGGGSDGGGGG----GGGGGGAEAEDGDGGGD 99
Score = 35.0 bits (79), Expect = 0.012
Identities = 20/45 (44%), Positives = 22/45 (48%)
Frame = -1
Query: 102 GGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GG G S GGG GGGGG GGG GGG + +SG
Sbjct: 206 GGGGRSESEGGGSDGGGGGG---GGGGGAEAEDGDGGGDLSSESG 81
Score = 31.6 bits (70), Expect = 0.13
Identities = 23/75 (30%), Positives = 31/75 (40%)
Frame = -1
Query: 61 GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG 120
G +K+ S +GGGG +D GGG G GGA GGG + G
Sbjct: 251 GRSKSRSSRLSLLRAGGGGRSESEGG---GSDGGGGGGGGGGGAEAEDGDGGGDLSSESG 81
Query: 121 GGEYSTGGGGGGYST 135
G ++ G G + T
Sbjct: 80 GALMTSAIGIGNWET 36
Score = 30.8 bits (68), Expect = 0.22
Identities = 17/36 (47%), Positives = 18/36 (49%), Gaps = 2/36 (5%)
Frame = -1
Query: 117 GGGGGGEYSTGG--GGGGYSTGGGGYGNDDSGNRRD 150
GGGG E GG GGGG GGGG +D D
Sbjct: 206 GGGGRSESEGGGSDGGGGGGGGGGGAEAEDGDGGGD 99
Score = 26.6 bits (57), Expect = 4.2
Identities = 34/139 (24%), Positives = 47/139 (33%), Gaps = 3/139 (2%)
Frame = +1
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DRP + Y++++ S P D D ++S S + ++ Y S
Sbjct: 244 DRPPNRRDYYDRSNNSPP-PKDRDRDFKRRRSPSPPSQRDRDRRHSPPPPYKRSRRGSPR 420
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEY---STGGGGGGYS 134
GGY Y D+ G Y GGY G GG Y + G G S
Sbjct: 421 GGY-------RYGPDDRFG-----------YDNSGGYERGMGGRVGYVDDKSYGRFGHRS 546
Query: 135 TGGGGYGNDDSGNRRDEVD 153
G G D + R D
Sbjct: 547 AGEYQNGISDVESNRGYAD 603
>AU251550
Length = 329
Score = 47.0 bits (110), Expect = 3e-06
Identities = 25/45 (55%), Positives = 26/45 (57%)
Frame = -3
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGY 140
GGG G GG GG GG G GGGGG GGGG G GGGG+
Sbjct: 327 GGGPGPGGCGG---CGGPGGPGGGGG**NMAGGGGDGGPCGGGGW 202
Score = 37.7 bits (86), Expect = 0.002
Identities = 26/55 (47%), Positives = 27/55 (48%), Gaps = 9/55 (16%)
Frame = -3
Query: 96 GGGYGTGGAGGG-----YSTGGGYGTGGGGGGEYSTGGGGGGY----STGGGGYG 141
GG G GG GGG + GGG G GGGG GGGGG S GG G G
Sbjct: 300 GGCGGPGGPGGGGG**NMAGGGGDGGPCGGGGW*CPNGGGGGAGA*GSRGGPGGG 136
Score = 37.0 bits (84), Expect = 0.003
Identities = 30/74 (40%), Positives = 32/74 (42%), Gaps = 6/74 (8%)
Frame = -3
Query: 74 NSGGGGYDSG-YNKTAYSTDEPSGGGYGTGGAGGGYSTGG-----GYGTGGGGGGEYSTG 127
N GGG D G + GGG G G+ GG GG G G GGGG G
Sbjct: 252 NMAGGGGDGGPCGGGGW*CPNGGGGGAGA*GSRGGPGGGGL**PGGCGGPGGGGLW*PGG 73
Query: 128 GGGGGYSTGGGGYG 141
GG GG GG G G
Sbjct: 72 GGPGG--PGGPGCG 37
Score = 34.3 bits (77), Expect = 0.020
Identities = 25/65 (38%), Positives = 29/65 (44%)
Frame = -3
Query: 73 PNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG 132
PN GGGG + ++ P GGG G GG GG + GGGG GG GG
Sbjct: 195 PNGGGGGAGA*GSRGG-----PGGGGL**PGGCGGPGGGGLW*PGGGG------PGGPGG 49
Query: 133 YSTGG 137
GG
Sbjct: 48 PGCGG 34
>TC19908 similar to GB|CAA60022.1|791150|VURNEXT26 extensin-like protein
{Vigna unguiculata;} , partial (49%)
Length = 472
Score = 45.1 bits (105), Expect = 1e-05
Identities = 28/59 (47%), Positives = 31/59 (52%), Gaps = 3/59 (5%)
Frame = -2
Query: 92 DEPSGGGYGTGGAGGGYSTGGGYGT---GGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
DE GGG GGG GGG GGGG GE GGGG Y+ GGGG G+ G+
Sbjct: 195 DEDGGGGDLYT*GGGGECEGGGGDL*I*GGGGDGE---GGGGDLYTYGGGGEGDGGGGD 28
Score = 43.5 bits (101), Expect = 3e-05
Identities = 22/36 (61%), Positives = 24/36 (66%)
Frame = -2
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGG 131
GGG G GG G Y+ GGG G G GGGG+ T GGGG
Sbjct: 108 GGGDGEGGGGDLYTYGGG-GEGDGGGGDL*T*GGGG 4
Score = 35.4 bits (80), Expect = 0.009
Identities = 23/55 (41%), Positives = 24/55 (42%), Gaps = 6/55 (10%)
Frame = -2
Query: 98 GYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGG------GGYGNDDSG 146
G G GG T GG G GGGG+ GGGG GG GG G D G
Sbjct: 201 GCDEDGGGGDLYT*GGGGECEGGGGDL*I*GGGGDGEGGGGDLYTYGGGGEGDGG 37
>TC16404 similar to UP|P93320 (P93320) Cdc2MsC protein, partial (15%)
Length = 584
Score = 45.1 bits (105), Expect = 1e-05
Identities = 31/83 (37%), Positives = 37/83 (44%), Gaps = 2/83 (2%)
Frame = +2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGG--YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGY 133
GGGGY SG P G G YG+ G GG GGG G G G GG GG
Sbjct: 23 GGGGYGSG-------PYPPQGRGAPYGSSGMPGGGPRGGGGGYGAGAPNYPQQGGPYGGS 181
Query: 134 STGGGGYGNDDSGNRRDEVDYKE 156
+ G G N GNR + +++
Sbjct: 182 AAGRG--SNMMGGNRNQQYGWQQ 244
Score = 36.6 bits (83), Expect = 0.004
Identities = 21/40 (52%), Positives = 23/40 (57%), Gaps = 5/40 (12%)
Frame = +2
Query: 107 GYSTGGGYGTGG----GGGGEY-STGGGGGGYSTGGGGYG 141
G GGGYG+G G G Y S+G GGG GGGGYG
Sbjct: 14 GXQGGGGYGSGPYPPQGRGAPYGSSGMPGGGPRGGGGGYG 133
>TC8694 similar to UP|IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c), partial (23%)
Length = 986
Score = 44.7 bits (104), Expect = 1e-05
Identities = 37/120 (30%), Positives = 48/120 (39%), Gaps = 13/120 (10%)
Frame = +2
Query: 43 DSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG----G 98
+S + + GGG + + +GGGG +G S +P G G
Sbjct: 389 ESNERAAEARIGGGGLDLPPRRRDGQDYAAAAAGGGG-GAGGRWQDLSLSQPRQGSGRAG 565
Query: 99 YGTGG--------AGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGY-GNDDSGNRR 149
YG+GG A GGYS GGGGGG S GG G GG G+ G+ R
Sbjct: 566 YGSGGGRPFAFNQAAGGYSRDRTGRGGGGGGGYQSQYGGSGRAHQGGSALRGHHGDGSSR 745
Score = 39.7 bits (91), Expect = 5e-04
Identities = 32/107 (29%), Positives = 39/107 (35%), Gaps = 17/107 (15%)
Frame = +2
Query: 72 EPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYST---------GGGYGTGGGGGG 122
E GGGG D + D + G GGAGG + G G G GGG
Sbjct: 410 EARIGGGGLDLPPRRRD-GQDYAAAAAGGGGGAGGRWQDLSLSQPRQGSGRAGYGSGGGR 586
Query: 123 EYSTGGGGGGYST--------GGGGYGNDDSGNRRDEVDYKEEEKHH 161
++ GGYS GGGGY + G+ R HH
Sbjct: 587 PFAFNQAAGGYSRDRTGRGGGGGGGYQSQYGGSGRAHQGGSALRGHH 727
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.303 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,433,566
Number of Sequences: 28460
Number of extensions: 75875
Number of successful extensions: 5487
Number of sequences better than 10.0: 582
Number of HSP's better than 10.0 without gapping: 1363
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3007
length of query: 243
length of database: 4,897,600
effective HSP length: 88
effective length of query: 155
effective length of database: 2,393,120
effective search space: 370933600
effective search space used: 370933600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0052b.4


