

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0050.4
(236 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
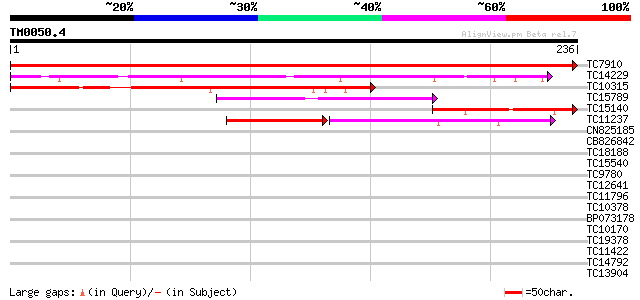
Score E
Sequences producing significant alignments: (bits) Value
TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%) 486 e-138
TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%) 183 2e-47
TC10315 similar to UP|P93166 (P93166) SCOF-1, partial (42%) 175 5e-45
TC15789 similar to UP|O22086 (O22086) ZPT2-14, partial (37%) 71 2e-13
TC15140 similar to UP|P93166 (P93166) SCOF-1, partial (25%) 64 2e-11
TC11237 similar to UP|Q43614 (Q43614) DNA-binding protein, parti... 57 3e-09
CN825185 34 0.019
CB826842 33 0.033
TC18188 similar to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6,... 33 0.043
TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fra... 33 0.056
TC9780 similar to AAQ56830 (AAQ56830) At5g09670, partial (55%) 32 0.096
TC12641 weakly similar to UP|Q9FM17 (Q9FM17) Similarity to nucle... 31 0.16
TC11796 similar to GB|AAP12870.1|30017273|BT006221 At4g35840 {Ar... 31 0.21
TC10378 similar to UP|Q84TG2 (Q84TG2) At3g16350, partial (26%) 30 0.37
BP073178 29 0.62
TC10170 similar to UP|PSD3_DAUCA (Q06364) Probable 26S proteasom... 29 0.62
TC19378 weakly similar to UP|O80967 (O80967) At2g39140 protein, ... 29 0.62
TC11422 UP|NIA_LOTJA (P39869) Nitrate reductase [NADH] (NR) , c... 29 0.81
TC14792 similar to UP|O73620 (O73620) Nuclear protein SDK2, part... 28 1.1
TC13904 weakly similar to UP|Q9ZU51 (Q9ZU51) RING-H2 finger prot... 28 1.8
>TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%)
Length = 1035
Score = 486 bits (1250), Expect = e-138
Identities = 236/236 (100%), Positives = 236/236 (100%)
Frame = +3
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 60
MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL
Sbjct: 93 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 272
Query: 61 CLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAA 120
CLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAA
Sbjct: 273 CLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAA 452
Query: 121 AEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATA 180
AEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATA
Sbjct: 453 AEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATA 632
Query: 181 SEGVGSTHSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPRLLPKIEIPHYY 236
SEGVGSTHSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPRLLPKIEIPHYY
Sbjct: 633 SEGVGSTHSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPRLLPKIEIPHYY 800
>TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%)
Length = 1175
Score = 183 bits (465), Expect = 2e-47
Identities = 123/281 (43%), Positives = 152/281 (53%), Gaps = 55/281 (19%)
Frame = +1
Query: 1 MAMEALKSPTTAAPTFTPF------QEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTE 54
MA+E L SPT A TPF +E PW K+KRSKR R D+ + TE
Sbjct: 55 MALETLNSPTAAT---TPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDN----PTTTE 213
Query: 55 EEYLALCLIMLARGST------------------AVTPKLTLSRPAPVTAEKLSYKCSVC 96
EEYLALCLIMLA+ + + + S P KL++KCSVC
Sbjct: 214 EEYLALCLIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVC 393
Query: 97 EKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSAS-----NGGGKVHQCSICQKS 151
+K FPSYQALGGHKASHRK +++E+ + ++A +AS GGK+H+CSIC KS
Sbjct: 394 DKAFPSYQALGGHKASHRK---SSSENQTPAAAAVNDNASVSTSTGNGGKMHECSICHKS 564
Query: 152 FPTGQALGGHKRCHYEGGGGASST-----------ATATASEGVGSTHSHQRNFDLNLPA 200
FPTGQALGGHKRCHYEGG ++ST T T+ G S HS R FDLNLPA
Sbjct: 565 FPTGQALGGHKRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAASAHS-LRGFDLNLPA 741
Query: 201 ------------FPDFSASKF-FVEEEVSSPLP--SKKPRL 226
D +K E+EV SPLP +K+PRL
Sbjct: 742 PLTEFWTPLGIGGGDSRTNKIRSGEQEVESPLPVTAKRPRL 864
>TC10315 similar to UP|P93166 (P93166) SCOF-1, partial (42%)
Length = 592
Score = 175 bits (444), Expect = 5e-45
Identities = 101/160 (63%), Positives = 112/160 (69%), Gaps = 8/160 (5%)
Frame = +2
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 60
MAMEAL SPTT AP+FT P + + PWAKRKRSKRSR SC+EEEYLAL
Sbjct: 140 MAMEALNSPTTTAPSFTFNDHPTLRHPAE-PWAKRKRSKRSR--------SCSEEEYLAL 292
Query: 61 CLIMLARGSTAVTPKLTLSRPA--PVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAG 118
CLIMLARG A T +PA P + +LSYKCSVC+K FPSYQALGGHKASHRK +G
Sbjct: 293 CLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSG 472
Query: 119 AAAEDHS---TSSAV--TTSSASNG-GGKVHQCSICQKSF 152
EDHS TSSAV TTSSA+ G GGK H+CSIC KSF
Sbjct: 473 GGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSF 592
>TC15789 similar to UP|O22086 (O22086) ZPT2-14, partial (37%)
Length = 747
Score = 70.9 bits (172), Expect = 2e-13
Identities = 36/92 (39%), Positives = 50/92 (54%)
Frame = +1
Query: 87 EKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCS 146
E + ++C C + F S+QALGGH+ASH E A T S N ++H CS
Sbjct: 229 ESVEFECKTCNRKFSSFQALGGHRASHNHKRVKLEEQ-----AKTPSLWDNNKPRMHVCS 393
Query: 147 ICQKSFPTGQALGGHKRCHYEGGGGASSTATA 178
+C F GQALGGH R H G +SS++++
Sbjct: 394 VCGLGFSLGQALGGHMRKHRNNEGFSSSSSSS 489
Score = 32.3 bits (72), Expect = 0.074
Identities = 16/36 (44%), Positives = 19/36 (52%)
Frame = +1
Query: 93 CSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSS 128
CSVC F QALGGH HR G ++ S+ S
Sbjct: 388 CSVCGLGFSLGQALGGHMRKHRNNEGFSSSSSSSYS 495
>TC15140 similar to UP|P93166 (P93166) SCOF-1, partial (25%)
Length = 503
Score = 63.9 bits (154), Expect = 2e-11
Identities = 36/68 (52%), Positives = 47/68 (68%), Gaps = 8/68 (11%)
Frame = +1
Query: 177 TATASEGVGSTH----SHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPR----LLP 228
+ATASEGVGSTH SH R+FDLN+PA P+F ++K E+EV SP P K + ++P
Sbjct: 1 SATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTK-AGEDEVESPHPVMKKKQRVFMIP 177
Query: 229 KIEIPHYY 236
KIEIP +
Sbjct: 178 KIEIPQLH 201
>TC11237 similar to UP|Q43614 (Q43614) DNA-binding protein, partial (20%)
Length = 523
Score = 57.0 bits (136), Expect = 3e-09
Identities = 36/98 (36%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Frame = +3
Query: 134 SASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTAT--ATASEGVGSTHSHQ 191
+ +N K+H+CSIC F +GQALGGH R H A+ T + T++ V +
Sbjct: 27 NGNNNKAKIHECSICGSEFTSGQALGGHMRRHRASANAAAETNSNAITSTTVVARPPRNI 206
Query: 192 RNFDLNLPAFP--DFSASKFFVEEEVSSPLPSKKPRLL 227
DLNLPA P D SKF S + S P L+
Sbjct: 207 LQLDLNLPAPPEDDLRESKFQFPATQKSMVLSAAPALV 320
Score = 44.3 bits (103), Expect = 2e-05
Identities = 21/42 (50%), Positives = 26/42 (61%)
Frame = +3
Query: 91 YKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTT 132
++CS+C F S QALGGH HR A AAAE +S + TT
Sbjct: 54 HECSICGSEFTSGQALGGHMRRHRASANAAAETNSNAITSTT 179
>CN825185
Length = 541
Score = 34.3 bits (77), Expect = 0.019
Identities = 17/39 (43%), Positives = 22/39 (55%)
Frame = +3
Query: 76 LTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHR 114
L+LS+P Y C C K F S QA+GGH+ +HR
Sbjct: 168 LSLSQPG-----SHGYGCLYCSKRFSSVQAVGGHQNAHR 269
Score = 30.8 bits (68), Expect = 0.21
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +3
Query: 134 SASNGGGKVHQCSICQKSFPTGQALGGHKRCH 165
S S G + C C K F + QA+GGH+ H
Sbjct: 171 SLSQPGSHGYGCLYCSKRFSSVQAVGGHQNAH 266
>CB826842
Length = 637
Score = 33.5 bits (75), Expect = 0.033
Identities = 18/48 (37%), Positives = 26/48 (53%), Gaps = 3/48 (6%)
Frame = +1
Query: 121 AEDHSTSSAVTTSSASNGG-GKV--HQCSICQKSFPTGQALGGHKRCH 165
A++ S S + S GG G+V + C+ C++ F QALGGH H
Sbjct: 52 AQNSSKSQQIGWRSDDLGGPGQVRSYSCTFCKRGFSNAQALGGHMNIH 195
Score = 33.1 bits (74), Expect = 0.043
Identities = 30/112 (26%), Positives = 45/112 (39%), Gaps = 3/112 (2%)
Frame = +1
Query: 90 SYKCSVCEKTFPSYQALGGHKASH---RKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCS 146
SY C+ C++ F + QALGGH H R ++E++ S ++ S + + +
Sbjct: 124 SYSCTFCKRGFSNAQALGGHMNIHTRDRAKLKQSSEENLLSLDISMKSTEHLDSEENLAP 303
Query: 147 ICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGVGSTHSHQRNFDLNL 198
QK H R S A AT S V S + DL+L
Sbjct: 304 --QKGKLVSATTAEHPR-------NPSYVAKATGSSVVVQMSSEEEKTDLDL 432
>TC18188 similar to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6, partial
(21%)
Length = 594
Score = 33.1 bits (74), Expect = 0.043
Identities = 15/49 (30%), Positives = 23/49 (46%), Gaps = 2/49 (4%)
Frame = +3
Query: 119 AAAEDHSTSSAVTTSSASN--GGGKVHQCSICQKSFPTGQALGGHKRCH 165
+ + D A ++S N G + +C C + F QALGGH+ H
Sbjct: 231 SGSSDSGNGDAAGSASGINTPSGDRKKKCQYCCREFANSQALGGHQNAH 377
Score = 32.0 bits (71), Expect = 0.096
Identities = 12/24 (50%), Positives = 17/24 (70%)
Frame = +3
Query: 92 KCSVCEKTFPSYQALGGHKASHRK 115
KC C + F + QALGGH+ +H+K
Sbjct: 312 KCQYCCREFANSQALGGHQNAHKK 383
>TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fragment),
partial (6%)
Length = 806
Score = 32.7 bits (73), Expect = 0.056
Identities = 28/97 (28%), Positives = 42/97 (42%), Gaps = 5/97 (5%)
Frame = +1
Query: 69 STAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSS 128
+ A T T + PAP T + S FP A ++ LA AAA S +
Sbjct: 25 AAATTTATTTTYPAPTTTAAAT-STSPTAAAFPDAAATTPSTSATTTLAAAAAATASAVA 201
Query: 129 AVTTS-----SASNGGGKVHQCSICQKSFPTGQALGG 160
A+T S +A++G G+ + SIC +S+ G
Sbjct: 202 AITASTTSSTAATDGWGR-NGSSICSRSWAVTNGFSG 309
>TC9780 similar to AAQ56830 (AAQ56830) At5g09670, partial (55%)
Length = 1250
Score = 32.0 bits (71), Expect = 0.096
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 4/39 (10%)
Frame = +2
Query: 136 SNGGGKVHQCSICQK----SFPTGQALGGHKRCHYEGGG 170
++GGGK + C K S P +A GG KRC Y GGG
Sbjct: 887 AHGGGKRCSFAGCSKGAEGSTPLCKAHGGGKRCLYNGGG 1003
>TC12641 weakly similar to UP|Q9FM17 (Q9FM17) Similarity to nuclear protein
ZAP, partial (3%)
Length = 396
Score = 31.2 bits (69), Expect = 0.16
Identities = 28/80 (35%), Positives = 37/80 (46%), Gaps = 5/80 (6%)
Frame = +3
Query: 110 KASHRKLAG--AAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYE 167
+ SHR L G +A H + TSS+ H S+ + P Q L C
Sbjct: 45 RPSHRVLLGPSSAFPPHPVTQPP*TSSS-------HPSSLRRSPSPPRQRLRSENPC--- 194
Query: 168 GGGGASSTATATA---SEGV 184
GGGG++S TATA +EGV
Sbjct: 195 GGGGSASVETATALPFAEGV 254
>TC11796 similar to GB|AAP12870.1|30017273|BT006221 At4g35840 {Arabidopsis
thaliana;}, partial (86%)
Length = 1114
Score = 30.8 bits (68), Expect = 0.21
Identities = 19/114 (16%), Positives = 43/114 (37%)
Frame = +1
Query: 53 TEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKAS 112
++E + L ++ + ++ +L R P + + E F Q +
Sbjct: 334 SDESGIGCLLYLIDVIGSLLSGRLVRERIGPAMLSAVQSQMGAVETGFDEVQNIFDTGCG 513
Query: 113 HRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHY 166
K + + +TT + ++ G+ CS+C + F G+ + CH+
Sbjct: 514 GAKGLSGDSVEKIPKIKITTDNNADASGERVSCSVCLQDFQLGETVRSLPHCHH 675
>TC10378 similar to UP|Q84TG2 (Q84TG2) At3g16350, partial (26%)
Length = 442
Score = 30.0 bits (66), Expect = 0.37
Identities = 20/63 (31%), Positives = 25/63 (38%)
Frame = +2
Query: 125 STSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGV 184
S+SS+ TT S G G +CS C K + C GGGG+ GV
Sbjct: 35 SSSSSTTTESRGGGIGMGRRCSHCSKENHNS------RTCPSRGGGGSGGGGGGVKLFGV 196
Query: 185 GST 187
T
Sbjct: 197 RLT 205
>BP073178
Length = 389
Score = 29.3 bits (64), Expect = 0.62
Identities = 19/54 (35%), Positives = 28/54 (51%)
Frame = -2
Query: 128 SAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATAS 181
SAVT + +S+GGG + S +G A GG GG +S++ T+T S
Sbjct: 355 SAVTAAGSSSGGGGGYNDSCPVSVANSGGAAGGGTGDTTPVGGDSSASLTSTTS 194
>TC10170 similar to UP|PSD3_DAUCA (Q06364) Probable 26S proteasome
non-ATPase regulatory subunit 3 (26S proteasome subunit
S3) (Nuclear antigen 21D7), partial (79%)
Length = 1197
Score = 29.3 bits (64), Expect = 0.62
Identities = 18/62 (29%), Positives = 27/62 (43%)
Frame = +3
Query: 33 AKRKRSKRSRTDSHHNHASCTEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYK 92
A++ RSK R ++H N C YL + + A L +R APV A +
Sbjct: 660 AEKLRSKAPRFEAHSNQQFCRYLFYLGKIRTIQLEYTDAKESLLQAARKAPVAARGFRIQ 839
Query: 93 CS 94
C+
Sbjct: 840 CN 845
>TC19378 weakly similar to UP|O80967 (O80967) At2g39140 protein, partial
(17%)
Length = 466
Score = 29.3 bits (64), Expect = 0.62
Identities = 22/66 (33%), Positives = 27/66 (40%), Gaps = 6/66 (9%)
Frame = +3
Query: 172 ASSTATATASEGVGSTHSHQRNFDLNLPAFPDF------SASKFFVEEEVSSPLPSKKPR 225
ASST TA T SH + + P S+SKF + S P P KP+
Sbjct: 33 ASSTMAMTALSLHCFTKSHSHSLTASFRTLPRLTCSFSSSSSKFNISFAPSKPKPKPKPK 212
Query: 226 LLPKIE 231
PK E
Sbjct: 213 PKPKPE 230
>TC11422 UP|NIA_LOTJA (P39869) Nitrate reductase [NADH] (NR) , complete
Length = 2932
Score = 28.9 bits (63), Expect = 0.81
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = +2
Query: 31 PWAKRKRSKRSRTDSHHNHASCT 53
PW+ + S S T SHH+HASCT
Sbjct: 272 PWSASQGSTLS-TRSHHSHASCT 337
>TC14792 similar to UP|O73620 (O73620) Nuclear protein SDK2, partial (5%)
Length = 492
Score = 28.5 bits (62), Expect = 1.1
Identities = 19/59 (32%), Positives = 26/59 (43%), Gaps = 6/59 (10%)
Frame = +3
Query: 8 SPTTAAPTF----TPFQEPNHSYMVDAPWAKRKRSKRSRTDS--HHNHASCTEEEYLAL 60
+PTT P+ ++P S PW K K K + S H+N+ EEE L L
Sbjct: 213 NPTTPWPSMKNQKNKIRKPKISLKSTTPWKKNKTQKHQQQQSLNHNNNEEEEEEEDLEL 389
>TC13904 weakly similar to UP|Q9ZU51 (Q9ZU51) RING-H2 finger protein RHA2b,
partial (32%)
Length = 535
Score = 27.7 bits (60), Expect = 1.8
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 7/39 (17%)
Frame = +1
Query: 137 NGGGKVHQCSICQKSFPTGQALGG-------HKRCHYEG 168
+G G+ H C +CQ++F G + H+RC +EG
Sbjct: 1 DGAGERHNCVVCQRTFKDGDQVRRLPCRHTFHRRC-FEG 114
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.126 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,790,103
Number of Sequences: 28460
Number of extensions: 74443
Number of successful extensions: 472
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 439
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 455
length of query: 236
length of database: 4,897,600
effective HSP length: 87
effective length of query: 149
effective length of database: 2,421,580
effective search space: 360815420
effective search space used: 360815420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0050.4


