

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.6
(1124 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
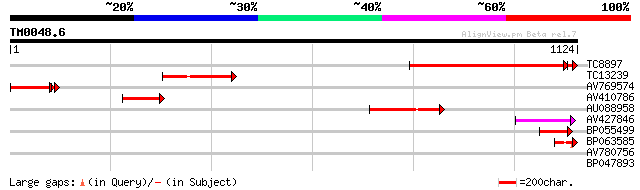
Score E
Sequences producing significant alignments: (bits) Value
TC8897 similar to UP|PHYA_PEA (P15001) Phytochrome A, partial (30%) 599 e-175
TC13239 homologue to UP|PHYB_SOYBN (P42499) Phytochrome B, parti... 201 7e-52
AV769574 167 2e-47
AV410786 175 3e-44
AU088958 169 2e-42
AV427846 83 3e-16
BP055499 60 3e-09
BP063585 49 6e-06
AV780756 35 0.051
BP047893 28 8.1
>TC8897 similar to UP|PHYA_PEA (P15001) Phytochrome A, partial (30%)
Length = 1250
Score = 599 bits (1544), Expect(2) = e-175
Identities = 313/316 (99%), Positives = 314/316 (99%)
Frame = +2
Query: 792 VMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGFFARSGKYVECL 851
VMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGFFARSGKYVECL
Sbjct: 2 VMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGFFARSGKYVECL 181
Query: 852 LSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALTYMKRQIRNPLS 911
LSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQ LSEQTALKRLKALTYMKRQIRNPLS
Sbjct: 182 LSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQRLSEQTALKRLKALTYMKRQIRNPLS 361
Query: 912 GIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYLDLEMAEFTLQD 971
GIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYLDLEMAEFTLQD
Sbjct: 362 GIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYLDLEMAEFTLQD 541
Query: 972 VLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLLISINCTPNGGQ 1031
VLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLLISINCTPNGGQ
Sbjct: 542 VLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLLISINCTPNGGQ 721
Query: 1032 VVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESEEGISLLISRKL 1091
VVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFG DGLESEEGISLLISRKL
Sbjct: 722 VVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGYDGLESEEGISLLISRKL 901
Query: 1092 LKLMSGDVRYLREAGK 1107
LKLMSGDVRYLREAG+
Sbjct: 902 LKLMSGDVRYLREAGQ 949
Score = 36.2 bits (82), Expect(2) = e-175
Identities = 18/18 (100%), Positives = 18/18 (100%)
Frame = +3
Query: 1107 KSSFILSVELAAAHKLKA 1124
KSSFILSVELAAAHKLKA
Sbjct: 948 KSSFILSVELAAAHKLKA 1001
>TC13239 homologue to UP|PHYB_SOYBN (P42499) Phytochrome B, partial (12%)
Length = 441
Score = 201 bits (510), Expect = 7e-52
Identities = 98/147 (66%), Positives = 115/147 (77%)
Frame = +3
Query: 303 DEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQKRK 362
DE L L L GSTLRAPH CH QYMANM SIASLVMAV++N ND+D G V +
Sbjct: 6 DEALMQPLCLVGSTLRAPHGCHAQYMANMGSIASLVMAVIINGNDDDAVG---VGGRSSM 176
Query: 363 RLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLCDM 422
RLWGLVVCH+TS R +PFPLRYACEFL Q F + +N E+++ Q LEK +LRTQTLLCDM
Sbjct: 177 RLWGLVVCHHTSARCIPFPLRYACEFLMQAFGLQLNMELQMAAQSLEKRVLRTQTLLCDM 356
Query: 423 LMRDAPLGILTQSPNLMDLVKCDGAAL 449
L+RD+P GI+TQSP++MDLVKCDGAAL
Sbjct: 357 LLRDSPAGIVTQSPSIMDLVKCDGAAL 437
>AV769574
Length = 540
Score = 167 bits (424), Expect(2) = 2e-47
Identities = 85/89 (95%), Positives = 85/89 (95%)
Frame = +1
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ
Sbjct: 241 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 420
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLAL 89
PKSNKVTTAYLHHIQRGKLIQPF L L
Sbjct: 421 PKSNKVTTAYLHHIQRGKLIQPFWLLAGL 507
Score = 39.7 bits (91), Expect(2) = 2e-47
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = +2
Query: 83 FGCLLALDEKTCKVIAY 99
FGCLLALDEKTCKVIAY
Sbjct: 488 FGCLLALDEKTCKVIAY 538
>AV410786
Length = 255
Score = 175 bits (444), Expect = 3e-44
Identities = 84/84 (100%), Positives = 84/84 (100%)
Frame = +2
Query: 224 DTMVQEVFELTGYDRVMAYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFM 283
DTMVQEVFELTGYDRVMAYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFM
Sbjct: 2 DTMVQEVFELTGYDRVMAYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFM 181
Query: 284 KNKVRMIVDCHAKQVKVLIDEKLP 307
KNKVRMIVDCHAKQVKVLIDEKLP
Sbjct: 182 KNKVRMIVDCHAKQVKVLIDEKLP 253
>AU088958
Length = 606
Score = 169 bits (428), Expect = 2e-42
Identities = 81/150 (54%), Positives = 104/150 (69%)
Frame = +3
Query: 713 NACASRDLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFG 772
NAC SRD +VGVCFV QDIT +K V+DKF ++EGDYKAI+Q+ NPLIPPIF +DE
Sbjct: 6 NACTSRDYTNAIVGVCFVGQDITHEKVVLDKFIKLEGDYKAIIQSLNPLIPPIFASDENA 185
Query: 773 WCCEWNPAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSE 832
C EWN AM KLTGWKREEV+ K+L GE+FG C+LK Q+ NF I+L ++G +
Sbjct: 186 CCSEWNAAMEKLTGWKREEVIGKLLPGEIFGNF---CQLKGQDTLTNFMILLYXGLSGQD 356
Query: 833 TEKVGFGFFARSGKYVECLLSVSKKLDVEG 862
+ K+ FGFF +GK+V ++ S K G
Sbjct: 357 SXKLPFGFFDXNGKFVXVYITASXKNXAAG 446
Score = 28.5 bits (62), Expect = 6.2
Identities = 16/52 (30%), Positives = 25/52 (47%)
Frame = +3
Query: 613 ELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAIGKHL 664
+LE +++ + PI A D + + WN + +LTG E IGK L
Sbjct: 105 KLEGDYKAIIQSLNPLIPPIFASDENACCSEWNAAMEKLTGWKREEVIGKLL 260
>AV427846
Length = 403
Score = 82.8 bits (203), Expect = 3e-16
Identities = 45/119 (37%), Positives = 70/119 (58%), Gaps = 1/119 (0%)
Frame = +1
Query: 1004 LYGDSLRLQQVLADFLLISINCTPN-GGQVVVAASLTKEQLGKSVHLANLELSITHGGSG 1062
+YGD LR+QQVLADFL + P+ G V + +Q+ + L + E + G G
Sbjct: 16 VYGDQLRIQQVLADFLSNVVRYAPSPDGWVEIHVYPKIKQISDGLTLLHAEFRLVCPGEG 195
Query: 1063 VPEALLNQMFGNDGLESEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
+P L+ MF N ++EG+ L +SRK+LKLM+G+V+Y+REA + F + +EL +
Sbjct: 196 LPSELVQDMFHNSRWVTQEGLGLCMSRKILKLMNGEVQYIREAERCYFFVLLELPVTRR 372
>BP055499
Length = 534
Score = 59.7 bits (143), Expect = 3e-09
Identities = 28/67 (41%), Positives = 46/67 (67%), Gaps = 1/67 (1%)
Frame = -3
Query: 1051 NLELSITHGGSGVPEALLNQMF-GNDGLESEEGISLLISRKLLKLMSGDVRYLREAGKSS 1109
+L+ +TH G G+P A+L+ MF G + ++EG+ L +SRK+L M+G V+Y+RE K
Sbjct: 481 HLKFRMTHSGQGLPSAVLHDMFEGGNQWTTQEGLGLYMSRKILSRMNGHVQYVREQNKCY 302
Query: 1110 FILSVEL 1116
F++ +EL
Sbjct: 301 FLIDLEL 281
>BP063585
Length = 382
Score = 48.5 bits (114), Expect = 6e-06
Identities = 33/48 (68%), Positives = 35/48 (72%), Gaps = 4/48 (8%)
Frame = -3
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFIL----SVELAAAHKLKA 1124
EGIS LI RKLLKLMSGDV R+A S I+ SVELAAAHKLKA
Sbjct: 380 EGISRLIRRKLLKLMSGDV---RDAKGSRPIMR*S*SVELAAAHKLKA 246
>AV780756
Length = 511
Score = 35.4 bits (80), Expect = 0.051
Identities = 18/37 (48%), Positives = 26/37 (69%), Gaps = 1/37 (2%)
Frame = -3
Query: 1082 GISLLISRKLLKLMSGDVRYL-REAGKSSFILSVELA 1117
G+ L +SRK+LKLM+G+V+Y RE SF++ LA
Sbjct: 509 GLGLCMSRKILKLMNGEVQYR*RERNAYSFVIQKRLA 399
>BP047893
Length = 514
Score = 28.1 bits (61), Expect = 8.1
Identities = 19/64 (29%), Positives = 27/64 (41%), Gaps = 5/64 (7%)
Frame = +2
Query: 320 PHSCHLQYMANMDSIASLVMAVVVND-----NDEDGDGSDSVQPQKRKRLWGLVVCHNTS 374
P SCH+ + DSI + V AVV + ND G + + R W + N S
Sbjct: 71 PSSCHIIMLRIFDSILARVKAVVTSPSSFHLNDPVGKKGGTNAKEATNRQWSAYMAVNCS 250
Query: 375 PRFV 378
+V
Sbjct: 251 KPWV 262
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,404,080
Number of Sequences: 28460
Number of extensions: 220736
Number of successful extensions: 1099
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1087
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1094
length of query: 1124
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1024
effective length of database: 2,051,600
effective search space: 2100838400
effective search space used: 2100838400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0048.6


