

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.13
(98 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
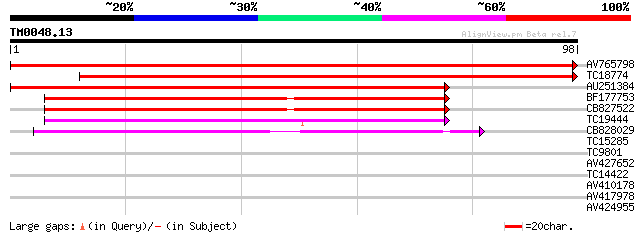
Score E
Sequences producing significant alignments: (bits) Value
AV765798 192 6e-51
TC18774 similar to GB|AAM16215.1|20334918|AY094059 At1g65700/F1E... 168 1e-43
AU251384 143 4e-36
BF177753 62 2e-11
CB827522 59 1e-10
TC19444 weakly similar to UP|LSM7_HUMAN (Q9UK45) U6 snRNA-associ... 48 3e-07
CB828029 47 5e-07
TC15285 homologue to PIR|T10586|T10586 small nuclear ribonucleop... 32 0.023
TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like... 25 2.2
AV427652 23 6.3
TC14422 similar to UP|Q9M5A8 (Q9M5A8) Chaperonin 21 precursor (1... 23 6.3
AV410178 23 6.3
AV417978 23 8.3
AV424955 23 8.3
>AV765798
Length = 566
Score = 192 bits (489), Expect = 6e-51
Identities = 98/98 (100%), Positives = 98/98 (100%)
Frame = +1
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALG 60
MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALG
Sbjct: 193 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALG 372
Query: 61 LYIIRGDNISVVGELDEDLDKNLDKSKIRAQPLKPVIH 98
LYIIRGDNISVVGELDEDLDKNLDKSKIRAQPLKPVIH
Sbjct: 373 LYIIRGDNISVVGELDEDLDKNLDKSKIRAQPLKPVIH 486
>TC18774 similar to GB|AAM16215.1|20334918|AY094059 At1g65700/F1E22_3
{Arabidopsis thaliana;}, partial (86%)
Length = 572
Score = 168 bits (426), Expect = 1e-43
Identities = 85/86 (98%), Positives = 86/86 (99%)
Frame = -1
Query: 13 QQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRGDNISVV 72
+QISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRGDNISVV
Sbjct: 461 EQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRGDNISVV 282
Query: 73 GELDEDLDKNLDKSKIRAQPLKPVIH 98
GELDEDLDKNLDKSKIRAQPLKPVIH
Sbjct: 281 GELDEDLDKNLDKSKIRAQPLKPVIH 204
>AU251384
Length = 364
Score = 143 bits (361), Expect = 4e-36
Identities = 73/76 (96%), Positives = 73/76 (96%)
Frame = +2
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALG 60
MSTGPGLESLVDQQISVITNDGRNIVG LKGFDQATNIILDESHERVFSTKEGVQ I LG
Sbjct: 137 MSTGPGLESLVDQQISVITNDGRNIVGVLKGFDQATNIILDESHERVFSTKEGVQQIVLG 316
Query: 61 LYIIRGDNISVVGELD 76
LYIIRGDNISVVGELD
Sbjct: 317 LYIIRGDNISVVGELD 364
>BF177753
Length = 491
Score = 61.6 bits (148), Expect = 2e-11
Identities = 31/70 (44%), Positives = 48/70 (68%)
Frame = +3
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L S +D+++ V+ DGR ++GTL+ FDQ N +L+ + ERV + I LGLY+IRG
Sbjct: 102 LASYLDKKLLVLLRDGRKLMGTLRSFDQFANAVLEGACERVI-VGDLYCDIPLGLYVIRG 278
Query: 67 DNISVVGELD 76
+N+ ++GELD
Sbjct: 279 ENVVLIGELD 308
>CB827522
Length = 519
Score = 59.3 bits (142), Expect = 1e-10
Identities = 28/70 (40%), Positives = 48/70 (68%)
Frame = +3
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L + +D+++ V+ DGR ++G L+ FDQ N++L+ + ERV + + LGLY+IRG
Sbjct: 177 LATYLDKKLLVLLRDGRKLLGLLRSFDQFANVVLEGACERVI-VGDLYCDVPLGLYVIRG 353
Query: 67 DNISVVGELD 76
+N+ ++GELD
Sbjct: 354 ENVVLIGELD 383
>TC19444 weakly similar to UP|LSM7_HUMAN (Q9UK45) U6 snRNA-associated
Sm-like protein LSm7, partial (88%)
Length = 565
Score = 47.8 bits (112), Expect = 3e-07
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 4/74 (5%)
Frame = +3
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFS----TKEGVQLIALGLY 62
L VD+ + V GR + GTLKG+DQ N++LDE+ E + K Q +LGL
Sbjct: 141 LAKFVDKGVQVKLTGGRQVTGTLKGYDQLLNLVLDEAVEFLRDPDDPLKTTAQTRSLGLI 320
Query: 63 IIRGDNISVVGELD 76
+ RG + +V D
Sbjct: 321 VCRGTAVMLVSPTD 362
>CB828029
Length = 502
Score = 47.0 bits (110), Expect = 5e-07
Identities = 26/78 (33%), Positives = 45/78 (57%)
Frame = +2
Query: 5 PGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
P L+ +D+ + + N R I+GTL+GFDQ N+++D + E G + +G+ +I
Sbjct: 77 PDLKKYMDKNLQIKLNANRMIIGTLRGFDQFMNLVIDNTVE-----VNGNEKNDIGMVVI 241
Query: 65 RGDNISVVGELDEDLDKN 82
RG+++ V L E + KN
Sbjct: 242 RGNSVVTVEAL-EPVTKN 292
>TC15285 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog
F9F13.90 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (61%)
Length = 587
Score = 31.6 bits (70), Expect = 0.023
Identities = 19/78 (24%), Positives = 35/78 (44%), Gaps = 9/78 (11%)
Frame = +3
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHE---------RVFSTK 51
MS + ++ ++ V DGR +VG FD+ N++L + E + S
Sbjct: 84 MSKSSKMLQYINYRMRVTIQDGRQLVGKFMAFDRHMNLVLGDCEEFRKLPPAKGKKPSDA 263
Query: 52 EGVQLIALGLYIIRGDNI 69
+ LGL ++RG+ +
Sbjct: 264 DREDRRTLGLVLLRGEEV 317
>TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like, complete
Length = 981
Score = 25.0 bits (53), Expect = 2.2
Identities = 21/75 (28%), Positives = 33/75 (44%)
Frame = +2
Query: 21 DGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRGDNISVVGELDEDLD 80
+G G L D NI L E + + K+G + + IRG+ I + DE +D
Sbjct: 158 NGETYNGHLVNCDTWMNIHL---REVICTNKDGDRFWRMPECYIRGNTIKYLRVPDEVID 328
Query: 81 KNLDKSKIRAQPLKP 95
K +++K R P
Sbjct: 329 KVQEETKSRTDRKPP 373
>AV427652
Length = 338
Score = 23.5 bits (49), Expect = 6.3
Identities = 9/26 (34%), Positives = 15/26 (57%)
Frame = +3
Query: 63 IIRGDNISVVGELDEDLDKNLDKSKI 88
+I GD+I GE ++ D+N D +
Sbjct: 228 VIAGDSIVQSGEASDECDENCDSHSL 305
>TC14422 similar to UP|Q9M5A8 (Q9M5A8) Chaperonin 21 precursor (10 kDa
chaperonin) (Protein Cpn10) (groES protein), partial
(93%)
Length = 1077
Score = 23.5 bits (49), Expect = 6.3
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Frame = +3
Query: 23 RNIVGTLKGFDQATNIILDESHERV-FSTKEGVQLIALGLYI 63
R ++ KGFDQ ++++LD S + GV L+ L L +
Sbjct: 165 RGMLHHSKGFDQFSSLVLDLSESPTPHGGRSGVCLLRLPLLL 290
>AV410178
Length = 430
Score = 23.5 bits (49), Expect = 6.3
Identities = 20/80 (25%), Positives = 35/80 (43%), Gaps = 2/80 (2%)
Frame = +1
Query: 20 NDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRGDNISVVGELDED- 78
+D RNI + + +I ++ + + E ++ L +N V E D +
Sbjct: 103 DDARNIGVVFNNIYELSGLIANDQYYSADTLSESQKVYVDTLVKKAYENWMHVIEYDGNS 282
Query: 79 -LDKNLDKSKIRAQPLKPVI 97
LD N +KS +QP PV+
Sbjct: 283 LLDYNQNKSLATSQPQAPVV 342
>AV417978
Length = 374
Score = 23.1 bits (48), Expect = 8.3
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = -1
Query: 37 NIILDESHERVFSTKEGVQL 56
N+ ++ESH RV KEG L
Sbjct: 80 NMAMEESH*RVLGEKEGFGL 21
>AV424955
Length = 320
Score = 23.1 bits (48), Expect = 8.3
Identities = 9/13 (69%), Positives = 11/13 (84%)
Frame = +1
Query: 38 IILDESHERVFST 50
IILDE+HER +T
Sbjct: 241 IILDEAHERTLAT 279
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.138 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,036,354
Number of Sequences: 28460
Number of extensions: 8498
Number of successful extensions: 40
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38
length of query: 98
length of database: 4,897,600
effective HSP length: 74
effective length of query: 24
effective length of database: 2,791,560
effective search space: 66997440
effective search space used: 66997440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 47 (22.7 bits)
Lotus: description of TM0048.13


