

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.12
(621 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
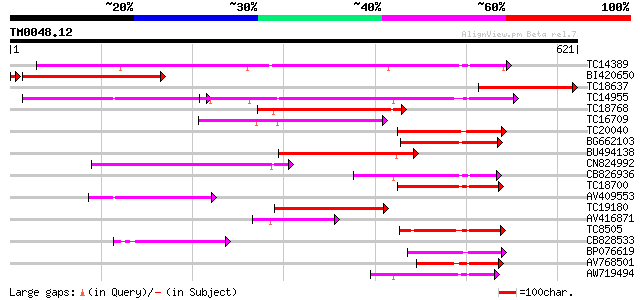
Score E
Sequences producing significant alignments: (bits) Value
TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 322 1e-88
BI420650 303 2e-85
TC18637 similar to UP|BAD12326 (BAD12326) NADH dehydrogenase sub... 210 6e-55
TC14955 UP|O22305 (O22305) Peptide transporter, complete 199 1e-51
TC18768 159 1e-39
TC16709 weakly similar to PIR|T47573|T47573 peptide transport-li... 149 1e-36
TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, ... 128 2e-30
BG662103 126 1e-29
BU494138 124 4e-29
CN824992 117 4e-27
CB826936 105 2e-23
TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like p... 101 3e-22
AV409553 94 5e-20
TC19180 similar to PIR|T10255|T10255 nitrite transport protein, ... 91 7e-19
AV416871 87 1e-17
TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partia... 86 2e-17
CB828533 80 1e-15
BP076619 78 4e-15
AV768501 76 1e-14
AW719494 72 3e-13
>TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(72%)
Length = 1990
Score = 322 bits (825), Expect = 1e-88
Identities = 189/540 (35%), Positives = 305/540 (56%), Gaps = 20/540 (3%)
Frame = +3
Query: 30 DNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVGGFISDTYLNRLTTCL 89
+ M + +LV++ +H +S+N ++N+ G + LVG +I+D YL R T +
Sbjct: 39 ERMAYYGISSNLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFI 218
Query: 90 LFGSLEVLALVMLTVQAGLDSLHPEACGKS----SCIKGG--IAVMLYTSLGLLALGLGG 143
+ + +L + +LT+ L +L P C + C + + Y +L ++ALG GG
Sbjct: 219 IASCIYLLGMCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGG 398
Query: 144 VRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTGVVWVNMQKGWHWGFS 203
+ + GADQFDE +P E +FFNW + S +G + T +V++ W G+
Sbjct: 399 TKPNISTMGADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYG 578
Query: 204 IITVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRNLPLPESNEQLY---- 259
+ TV + L+ +G PFYR K P SP+ R++QV V A +P +QL+
Sbjct: 579 LPTVGLAFSILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSM 758
Query: 260 EAYKDATVEKIAHTNQMRFLDKATILQENFEPRPWKVCTVTQVEEVKILTRMLPILASTI 319
E Y + + +I HT+ +RFLDKA + + + PW++CTVTQVEE K +T+M+PIL +T+
Sbjct: 759 EEYANNSRNRIDHTSTLRFLDKAAV--KTGKTSPWRLCTVTQVEETKQMTKMVPILITTL 932
Query: 320 IMNTCLAQLQTFSVSQGSVMSKKLG-SFEVPSPSIPVIPLFFLCVLIPIYEFIFVPFARK 378
I T L Q T + QG+ + + +G +FE+P + I + + IPIY+ +FVP R+
Sbjct: 933 IPCTMLIQAHTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRR 1112
Query: 379 ITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKR----RDQAI--KDPSKPISLFWLSF 432
T +P G+T LQR+G GL++ + M +A + E KR R++ + + P+S+F L
Sbjct: 1113YTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIP 1292
Query: 433 QYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVINTVTKR 492
QYA+ GVA+ F I ++FFY +AP MKSL S++ S++LG FLS+ ++S + +TK+
Sbjct: 1293QYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKK 1472
Query: 493 ITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYK---SEDNSCSKLN 549
S QGW+ +LN + L+ +Y F+ ILS LNF FL A + Y + + S S+LN
Sbjct: 1473--HSHQGWIL-DNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVTHEKSGSELN 1643
>BI420650
Length = 576
Score = 303 bits (777), Expect(2) = 2e-85
Identities = 156/156 (100%), Positives = 156/156 (100%)
Frame = +1
Query: 15 GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVG 74
GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVG
Sbjct: 109 GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVG 288
Query: 75 GFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIAVMLYTSL 134
GFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIAVMLYTSL
Sbjct: 289 GFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIAVMLYTSL 468
Query: 135 GLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATF 170
GLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATF
Sbjct: 469 GLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATF 576
Score = 30.4 bits (67), Expect(2) = 2e-85
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +2
Query: 1 MGDKEVKEWKRK 12
MGDKEVKEWKRK
Sbjct: 68 MGDKEVKEWKRK 103
>TC18637 similar to UP|BAD12326 (BAD12326) NADH dehydrogenase subunit 2,
partial (5%)
Length = 506
Score = 210 bits (534), Expect = 6e-55
Identities = 99/108 (91%), Positives = 105/108 (96%)
Frame = +1
Query: 514 FYWFLAILSCLNFFNFLYWASCYKYKSEDNSCSKLNVKAVAETTAIMVVDEKKHDKEDMK 573
FYWFLAILSCLNFF+FLYWASCY+Y+SED+SCSKLNVKAVAETTAIMVVDEKKHDKEDMK
Sbjct: 1 FYWFLAILSCLNFFDFLYWASCYQYQSEDHSCSKLNVKAVAETTAIMVVDEKKHDKEDMK 180
Query: 574 DHGSTQDLRAKAKESNQTSEANTEGPSSSDETDDGKQKERNSREWKDR 621
DHGST DLRAKAKESN TSEA+TEGPSSSDETDDG+ KERNSREWKDR
Sbjct: 181 DHGSTPDLRAKAKESNPTSEAHTEGPSSSDETDDGQPKERNSREWKDR 324
>TC14955 UP|O22305 (O22305) Peptide transporter, complete
Length = 2300
Score = 199 bits (506), Expect = 1e-51
Identities = 128/366 (34%), Positives = 205/366 (55%), Gaps = 17/366 (4%)
Frame = +1
Query: 209 SSIGFLILAL----GKPFYRIKA-PGDSPILRIVQVIVVAFKNRNLPLPESNEQLYEA-- 261
S+IGFL+ ++ G P YR K+ SP + ++V +VAF+NR L LP + +L+E
Sbjct: 682 SAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSELHEFQL 861
Query: 262 --YKDATVEKIAHTNQMRFLDKATILQENFEPRPWKVCTVTQVEEVKILTRMLPILASTI 319
Y + KI HT+ FLD+A I + N + CTVTQVE K++ M I +
Sbjct: 862 NYYISSGARKIHHTSHFSFLDRAAIRESNTDLSN-PPCTVTQVEGTKLVLGMFQIWLLML 1038
Query: 320 IMNTCLAQLQTFSVSQGSVMSKKLG-SFEVPSPSIPVIPLFFLCVLIPIYEFIFVPFARK 378
I C A T V QG+ M + LG F +P+ S+ + + +PIY+ F+PF R+
Sbjct: 1039 IPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMRR 1218
Query: 379 ITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAIK------DPSKPISLFWLSF 432
T + G+ LQRVG+G+ + I+MAV +E +R K + + P+S+FWL
Sbjct: 1219 RTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLLP 1398
Query: 433 QYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVINTVTKR 492
Q I GV++ F G+LEFFY ++P MK L T+ ++ G +++T +V++I+
Sbjct: 1399 QNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMID----- 1563
Query: 493 ITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYKSEDNSCSKL-NVK 551
K W+ G++LN ++L+ +Y FL ++S LNF FL+ +S Y YK E+ S +++ +++
Sbjct: 1564 ----KLNWI-GNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTSTTEVHDIE 1728
Query: 552 AVAETT 557
AE T
Sbjct: 1729 MSAEKT 1746
Score = 94.7 bits (234), Expect = 4e-20
Identities = 61/209 (29%), Positives = 96/209 (45%), Gaps = 2/209 (0%)
Frame = +3
Query: 15 GGFRASMFVFALSALDNMGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVG 74
G +A ++ L+ + +LV + ++ D+ SS + N+ G L ++G
Sbjct: 108 GKQKACTYILVYRVLERFAYYGVGANLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILG 287
Query: 75 GFISDTYLNRLTTCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGI--AVMLYT 132
+I+DTY R T + + LV+L + L SL P AC C + + YT
Sbjct: 288 AYIADTYTGRFWTITFSLLIYAIGLVLLVLTTTLKSLRP-ACENGICREASNLQVALFYT 464
Query: 133 SLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTGVVWV 192
SL +A+G G V+ M FGADQFD+ E + +FFNW + GS++ VV++
Sbjct: 465 SLYTIAVGSGAVKPNMSTFGADQFDDFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYI 644
Query: 193 NMQKGWHWGFSIITVASSIGFLILALGKP 221
+ G +G I S L LG P
Sbjct: 645 QEKNGXGFGLQFICYWFSTFLYYLFLGFP 731
>TC18768
Length = 738
Score = 159 bits (402), Expect = 1e-39
Identities = 81/168 (48%), Positives = 117/168 (69%), Gaps = 5/168 (2%)
Frame = +2
Query: 272 HTNQMRFLDKATILQE----NFEPRPWKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQ 327
HT++ RFLDKA I E N + W++C+V QVE+VKIL ++PI + TI+ NT LAQ
Sbjct: 236 HTDKFRFLDKACIRVEEAGSNTKKSSWRLCSVGQVEQVKILLSVIPIFSCTIVFNTILAQ 415
Query: 328 LQTFSVSQGSVMSKKLG-SFEVPSPSIPVIPLFFLCVLIPIYEFIFVPFARKITNHPSGV 386
LQTFSV QGS M L SF +P S+ IP L +++P+Y+ FVPFAR+IT H SG+
Sbjct: 416 LQTFSVQQGSAMDTHLTKSFHIPPASLQSIPYILLIIVVPLYDTFFVPFARRITGHESGI 595
Query: 387 TQLQRVGVGLVLSSISMAVAGIIEVKRRDQAIKDPSKPISLFWLSFQY 434
+ L+R+G GL L++ SM A ++E KRRD+A+ + +K +S+FW++ Q+
Sbjct: 596 SPLRRIGFGLFLATFSMVSAALLEKKRRDEAL-NHNKTLSIFWITPQF 736
>TC16709 weakly similar to PIR|T47573|T47573 peptide transport-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(41%)
Length = 712
Score = 149 bits (377), Expect = 1e-36
Identities = 78/217 (35%), Positives = 130/217 (58%), Gaps = 10/217 (4%)
Frame = +1
Query: 207 VASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRNLPLPESNEQLYE-AYKDA 265
VA ++ + G YR + PG SP+ R+ QV+V + + + P+ LYE A ++
Sbjct: 7 VAMAVAVVSFFSGTRLYRNQKPGGSPLTRMCQVVVASMRKCGVQAPDDKSLLYEIADTES 186
Query: 266 TVE---KIAHTNQMRFLDKATILQENFEPR----PWKVCTVTQVEEVKILTRMLPILAST 318
++ K+ HTN++ F DKA ++ ++ + + PW++CT TQVEE+K + RMLP+ A+
Sbjct: 187 AIKGSRKLDHTNELSFFDKAAVVVQSSDQKDSVDPWRLCTETQVEELKSILRMLPVWATG 366
Query: 319 IIMNTCLAQLQTFSVSQGSVMSKKLGS--FEVPSPSIPVIPLFFLCVLIPIYEFIFVPFA 376
II T Q+ T V QG M+ +G+ F++P ++ + + +P+Y++I VP A
Sbjct: 367 IIFATVYGQMSTLFVLQGQTMNTHVGNSNFKIPPAALSIFDTLSVIFWVPVYDWIIVPIA 546
Query: 377 RKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKR 413
RK + H +G+TQLQR+G+GL +S +M A I+EV R
Sbjct: 547 RKFSGHKNGLTQLQRMGIGLFISIFAMVAAAILEVIR 657
>TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, partial
(20%)
Length = 578
Score = 128 bits (322), Expect = 2e-30
Identities = 60/122 (49%), Positives = 87/122 (71%), Gaps = 2/122 (1%)
Frame = +3
Query: 425 ISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVS 484
++ W++FQY G AD+FTL G++EFF+ EAP +M+SL+T+ ++ S+S+GYFLSTV+VS
Sbjct: 3 MTFLWVAFQYLFLGSADLFTLAGMMEFFFTEAPWSMRSLATALSWASLSMGYFLSTVLVS 182
Query: 485 VINTVTKRI--TPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYKSED 542
+IN VT TP WL GS+LN +L FYW + +LS LNF ++L+WA+ YKY+
Sbjct: 183 IINKVTGAFGHTP----WLLGSNLNHYHLERFYWLMCVLSGLNFIHYLFWANSYKYRGSP 350
Query: 543 NS 544
S
Sbjct: 351 RS 356
>BG662103
Length = 387
Score = 126 bits (316), Expect = 1e-29
Identities = 56/111 (50%), Positives = 84/111 (75%)
Frame = +1
Query: 429 WLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVINT 488
W++FQY G AD+FTL GLLEFF+ EAP M+SL+TS ++ S+++GY+LS+ +VS++N+
Sbjct: 19 WIAFQYLFLGSADLFTLAGLLEFFFSEAPIRMRSLATSLSWASLAIGYYLSSAIVSIVNS 198
Query: 489 VTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYK 539
VT + S + WL G++LN +L FYW + +LS LNF ++LYWA+ YKY+
Sbjct: 199 VTGK--GSHKPWLSGANLNHYHLERFYWLMCLLSGLNFLHYLYWAARYKYR 345
>BU494138
Length = 518
Score = 124 bits (311), Expect = 4e-29
Identities = 65/158 (41%), Positives = 100/158 (63%), Gaps = 5/158 (3%)
Frame = +2
Query: 295 KVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLGS-FEVPSPSI 353
+V ++ QVEE+K L R+ PI A+ I+ T +AQ TF+VSQ M + +GS F++P+ S+
Sbjct: 11 RVVSIQQVEEIKCLARIFPIWAAGILGFTAMAQQGTFTVSQAMKMDRHIGSKFQIPAGSL 190
Query: 354 PVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKR 413
VI + + +P Y+ FVP R+IT H G+T LQR+G+G+V S +SM VAG++E R
Sbjct: 191 GVISFITIGLWVPFYDRFFVPAVRRITKHEGGITLLQRIGIGMVFSVLSMIVAGLVEKVR 370
Query: 414 RDQAIKDPS----KPISLFWLSFQYAIFGVADMFTLIG 447
R A +P+ P+S+ WL+ Q + G+ + F IG
Sbjct: 371 RGVANSNPNPLGIAPMSVMWLAPQLVLMGLCEAFNAIG 484
>CN824992
Length = 708
Score = 117 bits (294), Expect = 4e-27
Identities = 80/235 (34%), Positives = 120/235 (51%), Gaps = 13/235 (5%)
Frame = +2
Query: 90 LFGSLEVLALVMLTVQAGLDSLHPEACGKSS--CIKGGI--AVMLYTSLGLLALGLGGVR 145
+F + V+ L L++ L L P CG C + Y S+ L+ALG GG +
Sbjct: 5 IFQVIFVVGLASLSLTTYLFLLEPNGCGDKELPCTSHSSYQTALFYVSIYLIALGNGGYQ 184
Query: 146 GAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTGVVWVNMQKGWHWGFSII 205
+ FGADQFDE+DP E + +FF++ L+ LGS+ T + + W GF
Sbjct: 185 PNIATFGADQFDEEDPKEQHSKISFFSYFYLALNLGSLFSNTILNYFEDDGLWTLGFWAS 364
Query: 206 TVASSIGFLILALGKPFYRIKAPGDSPILRIVQVIVVAFKNRNLPL-PESNEQLYEAYKD 264
++ + ++ G P+YR PG +P+ R QV V A + + + P +QL+E +
Sbjct: 365 AGSAFLALVLFLCGTPWYRYFKPGGNPVPRFCQVFVAAMRKWKVKVSPGEEDQLHEVEEC 544
Query: 265 AT--VEKIAHTNQMRFLDKATIL------QENFEPRPWKVCTVTQVEEVKILTRM 311
+T K+ HT RFLDKA ++ QEN E W + TVTQVEEVK + R+
Sbjct: 545 STNGGRKMFHTEGFRFLDKAALITSQESKQEN-ECSLWHLSTVTQVEEVKCILRL 706
>CB826936
Length = 596
Score = 105 bits (262), Expect = 2e-23
Identities = 64/168 (38%), Positives = 99/168 (58%), Gaps = 6/168 (3%)
Frame = +3
Query: 377 RKITNHPSGVTQLQRVGVGLVLSSISMAVAGIIEVKRRDQAIK------DPSKPISLFWL 430
R+ T +P G+ LQR+G GLV+ I+M A + E KR A + P+++F L
Sbjct: 9 RQYTKNPRGIAMLQRLGAGLVMYIITMVTAWLSERKRLSVAREYNLLGQHDKIPLTIFIL 188
Query: 431 SFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVINTVT 490
Q+A+ GVA+ F I +L+FFY +AP MKSL S++ S +LG F S+ ++S + +T
Sbjct: 189 LPQFALTGVANNFVDIAMLDFFYDQAPEGMKSLGISYSTTSTALGCFFSSFLLSTVADLT 368
Query: 491 KRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKY 538
K+ +GW+ +LN ++L+ +Y F+AILS NF FL A + Y
Sbjct: 369 KK--HGHKGWVL-DNLNVSHLDYYYAFMAILSVANFLCFLVAAKLFAY 503
>TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like protein,
partial (18%)
Length = 615
Score = 101 bits (252), Expect = 3e-22
Identities = 47/117 (40%), Positives = 75/117 (63%)
Frame = +1
Query: 425 ISLFWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVS 484
+S +WL QY + GVA++F ++GLLEF Y EAP MKS+ +++ L+ LG F +T++ S
Sbjct: 46 LSAYWLLIQYCLIGVAEVFCIVGLLEFLYEEAPDAMKSIGSAYAALAGGLGCFAATIINS 225
Query: 485 VINTVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYKSE 541
+I ++T + K+ WL ++N + FYW L LS +NF F+Y A YKY+++
Sbjct: 226 IIKSLTGK--EGKESWL-AQNINTGRFDYFYWILTALSLVNFCIFIYSAHRYKYRTQ 387
>AV409553
Length = 433
Score = 94.4 bits (233), Expect = 5e-20
Identities = 51/142 (35%), Positives = 77/142 (53%), Gaps = 2/142 (1%)
Frame = +2
Query: 87 TCLLFGSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIA--VMLYTSLGLLALGLGGV 144
T L + V+ +++LTV L SL P C C K + V YT+L A+G GG
Sbjct: 2 TFTLSSIVYVMGMILLTVAVSLKSLKP-TCTNGICNKASTSQIVFFYTALYTTAIGAGGT 178
Query: 145 RGAMVAFGADQFDEKDPVEAKALATFFNWLLLSSTLGSVVGVTGVVWVNMQKGWHWGFSI 204
+ + FGADQFD+ +P E + A+FFNW + +S LG+++ G+V++ GW G+ I
Sbjct: 179 KPNISTFGADQFDDFNPHEKQTKASFFNWWMFTSFLGALIATLGLVYIQENLGWGLGYGI 358
Query: 205 ITVASSIGFLILALGKPFYRIK 226
T + +I +G P YR K
Sbjct: 359 PTSGLLLSLVIFYVGTPMYRHK 424
>TC19180 similar to PIR|T10255|T10255 nitrite transport protein, chloroplast
- cucumber {Cucumis sativus;} , partial (12%)
Length = 408
Score = 90.5 bits (223), Expect = 7e-19
Identities = 46/126 (36%), Positives = 78/126 (61%), Gaps = 1/126 (0%)
Frame = +1
Query: 291 PRPWKVCTVTQVEEVKILTRMLPILASTIIMNTCLAQLQTFSVSQGSVMSKKLG-SFEVP 349
P W++ TV +VEE+K + RM PI AS I++ T AQ TFS+ Q M + + SF++P
Sbjct: 31 PNKWRLNTVHRVEELKSIIRMGPIWASGILLITAYAQQGTFSLQQAKTMDRHITKSFQIP 210
Query: 350 SPSIPVIPLFFLCVLIPIYEFIFVPFARKITNHPSGVTQLQRVGVGLVLSSISMAVAGII 409
+ S+ V + + +Y+ + + AR+ T G++ L R+G+G V+S+I+ VAG +
Sbjct: 211 AGSMSVFTIITMLTTTALYDRVLIRVARRFTGLDRGISFLHRMGIGFVISTIATFVAGFV 390
Query: 410 EVKRRD 415
E+KR++
Sbjct: 391 EMKRKN 408
>AV416871
Length = 354
Score = 86.7 bits (213), Expect = 1e-17
Identities = 47/103 (45%), Positives = 61/103 (58%), Gaps = 8/103 (7%)
Frame = +3
Query: 267 VEKIAHTNQMRFLDKATI-------LQENFEPRPWKVCTVTQVEEVKILTRMLPILASTI 319
+E + H + RFLDKA I L++ WK+C VTQVE KI+ M P+ TI
Sbjct: 39 IEFLPHRDIFRFLDKAAIQRECDMELEKPESTSQWKLCRVTQVENAKIILSMGPVFLCTI 218
Query: 320 IMNTCLAQLQTFSVSQGSVMSKKL-GSFEVPSPSIPVIPLFFL 361
IM CLAQ+QTFSV QG M K+ F +P S+P+IP+ FL
Sbjct: 219 IMTLCLAQIQTFSVQQGYTMDTKITKDFNMPPASLPIIPVIFL 347
>TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partial (23%)
Length = 1495
Score = 85.5 bits (210), Expect = 2e-17
Identities = 49/116 (42%), Positives = 73/116 (62%)
Frame = +3
Query: 428 FWLSFQYAIFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVIN 487
++LSF + G A F +G LEFF REAP MKS+ST ++S+GYF+S+++VS+++
Sbjct: 210 WFLSFFWWEQGKA--FAYVGQLEFFIREAPERMKSMSTGLFLAALSMGYFVSSLLVSIVD 383
Query: 488 TVTKRITPSKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYKSEDN 543
+ SK+ WL S+LNK L+ FYW LA+L LNF F+ A + Y+ + N
Sbjct: 384 KL------SKKKWL-KSNLNKGRLDYFYWLLAVLGVLNFILFIVLAMRHHYQVQHN 530
>CB828533
Length = 557
Score = 80.1 bits (196), Expect = 1e-15
Identities = 47/128 (36%), Positives = 66/128 (50%)
Frame = +1
Query: 114 EACGKSSCIKGGIAVMLYTSLGLLALGLGGVRGAMVAFGADQFDEKDPVEAKALATFFNW 173
EA GK + + V LYT +A+G GG++ ++ FG+DQFD DP E K + FFN
Sbjct: 193 EASGKQVAL---LLVALYT----IAVGAGGIKSSVSGFGSDQFDTTDPREEKKMVFFFNR 351
Query: 174 LLLSSTLGSVVGVTGVVWVNMQKGWHWGFSIITVASSIGFLILALGKPFYRIKAPGDSPI 233
+ GS+ V +V+V G WGF I + L GKP YR K P SP+
Sbjct: 352 FYFFVSTGSLFSVLVLVYVQDNIGRGWGFGIPAGIMLVSLAFLLYGKPLYRYKRPQGSPL 531
Query: 234 LRIVQVIV 241
I +V++
Sbjct: 532 TLIWKVLI 555
>BP076619
Length = 432
Score = 78.2 bits (191), Expect = 4e-15
Identities = 41/109 (37%), Positives = 66/109 (59%)
Frame = -3
Query: 436 IFGVADMFTLIGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVINTVTKRITP 495
+FGV+++FTL+GL EFFY + P ++SL + +G FLS ++SVI TVT + P
Sbjct: 427 LFGVSEVFTLVGLQEFFYDQVPNELRSLGLALYLSIFGVGSFLSGFLISVIETVTGKDGP 248
Query: 496 SKQGWLYGSDLNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYKSEDNS 544
+ W + ++L+K +++ FYW LA S + F F+ +A Y Y + S
Sbjct: 247 DR--W-FANNLHKAHIDYFYWLLAGFSVVGFAMFMCFAKSYVYNHQGAS 110
>AV768501
Length = 442
Score = 76.3 bits (186), Expect = 1e-14
Identities = 39/96 (40%), Positives = 62/96 (63%)
Frame = -1
Query: 446 IGLLEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSD 505
IG L+FF RE P MK++S ++SLG+F S+++V++++ ++T +QGWL +
Sbjct: 442 IGQLDFFLRECPKGMKTMSPGLFLSTLSLGFFFSSMLVTLVH----KMTGPRQGWL-ADN 278
Query: 506 LNKNNLNLFYWFLAILSCLNFFNFLYWASCYKYKSE 541
LN+ L+ FYW LA+LS +N FL+ A Y YK +
Sbjct: 277 LNQGKLDDFYWLLAVLSGVNLVVFLFCAKWYVYKDK 170
>AW719494
Length = 473
Score = 71.6 bits (174), Expect = 3e-13
Identities = 43/148 (29%), Positives = 75/148 (50%), Gaps = 7/148 (4%)
Frame = +3
Query: 396 LVLSSISMAVAGIIEVKRRDQAIK-------DPSKPISLFWLSFQYAIFGVADMFTLIGL 448
L+ S + + + +E RR +AI+ +S WL+ Q + G+A+ F IG
Sbjct: 3 LLFSFLHLVTSATLETIRRKRAIEAGYINNAHGVLNMSAMWLAPQLCLGGIAEAFNAIGQ 182
Query: 449 LEFFYREAPPTMKSLSTSFTYLSMSLGYFLSTVVVSVINTVTKRITPSKQGWLYGSDLNK 508
EF+Y E P TM +++S L M+ G +S+ V S + +T + K+GW+ ++NK
Sbjct: 183 NEFYYTEFPRTMSXVASSLLGLGMAAGNIVSSFVFSAVENITS--SGGKEGWV-SDNINK 353
Query: 509 NNLNLFYWFLAILSCLNFFNFLYWASCY 536
+ +YW + +S LN +L + Y
Sbjct: 354 GRFDKYYWVVXGISGLNLVYYLICSWAY 437
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,912,344
Number of Sequences: 28460
Number of extensions: 155915
Number of successful extensions: 1053
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 1014
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1019
length of query: 621
length of database: 4,897,600
effective HSP length: 96
effective length of query: 525
effective length of database: 2,165,440
effective search space: 1136856000
effective search space used: 1136856000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0048.12


