

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0042.2
(173 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
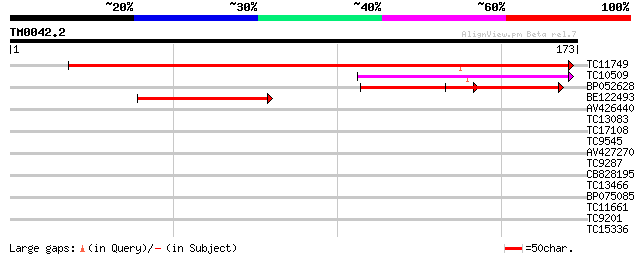
TC11749 weakly similar to UP|Q9ZVL7 (Q9ZVL7) T22H22.18, partial ... 190 1e-49
TC10509 weakly similar to UP|Q7PWW3 (Q7PWW3) ENSANGP00000016794 ... 96 3e-21
BP052628 69 3e-13
BE122493 54 2e-08
AV426440 28 0.86
TC13083 27 1.9
TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, pa... 27 2.5
TC9545 similar to UP|Q9XGC3 (Q9XGC3) SINA2p, partial (40%) 26 3.3
AV427270 26 3.3
TC9287 25 5.6
CB828195 25 5.6
TC13466 weakly similar to UP|Q9FK19 (Q9FK19) Genomic DNA, chromo... 25 7.3
BP075085 25 7.3
TC11661 similar to UP|Q93ZF7 (Q93ZF7) AT4g33140/F4I10_70, partia... 25 9.5
TC9201 25 9.5
TC15336 25 9.5
>TC11749 weakly similar to UP|Q9ZVL7 (Q9ZVL7) T22H22.18, partial (28%)
Length = 948
Score = 190 bits (483), Expect = 1e-49
Identities = 102/189 (53%), Positives = 119/189 (61%), Gaps = 35/189 (18%)
Frame = +1
Query: 19 LRWSQRRHFCAVLCRDDGSHSTSQAESSTITLWKHNSELVHALFVPPNEPTKLNKLLREQ 78
L W + + L DGS ++ E+S+ T+WK NSELV LFVPPN+P KLNKLLR+Q
Sbjct: 106 LTWQPQLPISSSLKATDGSQIKTRTEASSSTVWKPNSELVDGLFVPPNDPRKLNKLLRKQ 285
Query: 79 VKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKKGNSKSKALPKYFQASS-- 136
VKDT G+ WF+MPAQTI PELQKD L+LR AIDPKRHYKK +SKSK LPKYFQ +
Sbjct: 286 VKDTAGKNWFNMPAQTITPELQKDLKLLKLRGAIDPKRHYKKCDSKSKTLPKYFQMGTVV 465
Query: 137 ---------------------------------GARKVREIEEQNQPAGNEKWKVKGGNS 163
RKVREIEE NQPAGNE WK+KGG+S
Sbjct: 466 DSPLDFFSGRLTKAERKASFAEELLSDPNLEAYRKRKVREIEEPNQPAGNENWKIKGGSS 645
Query: 164 RKRAKERRN 172
RKRAKE+RN
Sbjct: 646 RKRAKEKRN 672
>TC10509 weakly similar to UP|Q7PWW3 (Q7PWW3) ENSANGP00000016794 (Fragment),
partial (16%)
Length = 548
Score = 95.9 bits (237), Expect = 3e-21
Identities = 52/101 (51%), Positives = 59/101 (57%), Gaps = 35/101 (34%)
Frame = +2
Query: 107 QLRAAIDPKRHYKKGNSKSKALPKYFQASSGA---------------------------- 138
QLR AIDPKRHYKKG+SKSK LP+YFQ +
Sbjct: 86 QLRGAIDPKRHYKKGDSKSKTLPQYFQMGTVVDSPLDFFSGRLTKKERRASLADELLADQ 265
Query: 139 -------RKVREIEEQNQPAGNEKWKVKGGNSRKRAKERRN 172
RKVREIEEQN+PAGNE WK+KGG+SRKRAKE+RN
Sbjct: 266 NLVAYRKRKVREIEEQNKPAGNENWKIKGGSSRKRAKEKRN 388
>BP052628
Length = 528
Score = 69.3 bits (168), Expect = 3e-13
Identities = 33/36 (91%), Positives = 34/36 (93%)
Frame = +3
Query: 134 ASSGARKVREIEEQNQPAGNEKWKVKGGNSRKRAKE 169
A+ ARKVREIEEQNQPAGNEKWKVKGGNSRKRAKE
Sbjct: 420 AAYRARKVREIEEQNQPAGNEKWKVKGGNSRKRAKE 527
Score = 61.2 bits (147), Expect = 9e-11
Identities = 29/36 (80%), Positives = 30/36 (82%)
Frame = +2
Query: 108 LRAAIDPKRHYKKGNSKSKALPKYFQASSGARKVRE 143
LRAAIDPKRHYKKGNSKSKALPKYFQ S K R+
Sbjct: 269 LRAAIDPKRHYKKGNSKSKALPKYFQVGSKLTKKRD 376
>BE122493
Length = 285
Score = 53.5 bits (127), Expect = 2e-08
Identities = 24/41 (58%), Positives = 30/41 (72%)
Frame = +2
Query: 40 TSQAESSTITLWKHNSELVHALFVPPNEPTKLNKLLREQVK 80
++Q E + T+WK NSEL+ LFVPPN P LN LLR+QVK
Sbjct: 140 STQTEPLSSTVWKSNSELIDGLFVPPNAPXNLNXLLRKQVK 262
>AV426440
Length = 264
Score = 28.1 bits (61), Expect = 0.86
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = -1
Query: 142 REIEEQNQPAGNEKWKVKGGNSRKRAKER 170
RE E QNQ + K +++GG R R++ER
Sbjct: 231 RE*ESQNQNQRSRKDELEGGRRRSRSRER 145
>TC13083
Length = 528
Score = 26.9 bits (58), Expect = 1.9
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = +3
Query: 101 KDETSLQLRAAIDPKRHYKKGNSKSK 126
KDE+S + +DPKR ++ SKS+
Sbjct: 204 KDESSSSCKVELDPKRIIRRARSKSE 281
>TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (16%)
Length = 573
Score = 26.6 bits (57), Expect = 2.5
Identities = 9/27 (33%), Positives = 17/27 (62%)
Frame = -2
Query: 17 TKLRWSQRRHFCAVLCRDDGSHSTSQA 43
TK++W + HFC++ ++ + ST A
Sbjct: 530 TKMKWCKYHHFCSLDALNESNESTRAA 450
>TC9545 similar to UP|Q9XGC3 (Q9XGC3) SINA2p, partial (40%)
Length = 543
Score = 26.2 bits (56), Expect = 3.3
Identities = 15/43 (34%), Positives = 21/43 (47%)
Frame = +2
Query: 20 RWSQRRHFCAVLCRDDGSHSTSQAESSTITLWKHNSELVHALF 62
+W Q A + R+ HS +ESS + W +NSE LF
Sbjct: 218 KWPQ-----AYMARNSEEHS*QSSESSR*SRWSYNSEEPWPLF 331
>AV427270
Length = 267
Score = 26.2 bits (56), Expect = 3.3
Identities = 16/55 (29%), Positives = 24/55 (43%)
Frame = +1
Query: 7 ERIRLRSENPTKLRWSQRRHFCAVLCRDDGSHSTSQAESSTITLWKHNSELVHAL 61
++I LRS +WS RRHF + G+ S + + LW S H +
Sbjct: 55 KKILLRS------KWSTRRHFWGTPVQSPGAASLHLETTLLVPLWMELSGYGHMM 201
>TC9287
Length = 566
Score = 25.4 bits (54), Expect = 5.6
Identities = 21/55 (38%), Positives = 25/55 (45%), Gaps = 1/55 (1%)
Frame = +1
Query: 4 QRTERIRLRSENPTKLRWSQRR-HFCAVLCRDDGSHSTSQAESSTITLWKHNSEL 57
QR E RSE + WS CA+ CR + Q S +IT WKH S L
Sbjct: 82 QRFEGRTTRSET-SGFEWSHLSIQ*CALHCRV--LNHNHQTTSPSITPWKHRSTL 237
>CB828195
Length = 551
Score = 25.4 bits (54), Expect = 5.6
Identities = 19/64 (29%), Positives = 26/64 (39%), Gaps = 15/64 (23%)
Frame = -2
Query: 88 FDMPAQTINPELQKDETSLQLRAAIDPKRHYKK---------------GNSKSKALPKYF 132
F MPA + NP+ Q+ T A+I+P K NS +L YF
Sbjct: 520 FSMPASSFNPKTQR--TLCSSLASINPSLFSSKTSKACFTSSKLGSWSSNSSESSLEPYF 347
Query: 133 QASS 136
+SS
Sbjct: 346 SSSS 335
>TC13466 weakly similar to UP|Q9FK19 (Q9FK19) Genomic DNA, chromosome 5, TAC
clone:K19E1, partial (16%)
Length = 552
Score = 25.0 bits (53), Expect = 7.3
Identities = 12/42 (28%), Positives = 19/42 (44%)
Frame = -2
Query: 56 ELVHALFVPPNEPTKLNKLLREQVKDTTGRIWFDMPAQTINP 97
+++H +VP PT +K L ++ G I M NP
Sbjct: 476 KMIHHYWVPSPRPTNKHKTLEMTCENLMGEIENKMRKDVFNP 351
>BP075085
Length = 407
Score = 25.0 bits (53), Expect = 7.3
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = -1
Query: 152 GNEKWKVKGGNSRK 165
GNE+WK GG S+K
Sbjct: 89 GNERWKGGGGKSKK 48
>TC11661 similar to UP|Q93ZF7 (Q93ZF7) AT4g33140/F4I10_70, partial (20%)
Length = 651
Score = 24.6 bits (52), Expect = 9.5
Identities = 10/23 (43%), Positives = 14/23 (60%)
Frame = -3
Query: 53 HNSELVHALFVPPNEPTKLNKLL 75
HN L+H + + PN PT+ LL
Sbjct: 520 HNQFLLHLIHI*PNNPTQKRILL 452
>TC9201
Length = 1171
Score = 24.6 bits (52), Expect = 9.5
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = -2
Query: 4 QRTERIRLRSENPTKLRWSQRRHF 27
Q ERIR++ E+ + WS+R+ F
Sbjct: 405 QPNERIRIQHESGAENGWSRRKKF 334
>TC15336
Length = 1145
Score = 24.6 bits (52), Expect = 9.5
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = -1
Query: 38 HSTSQAESSTITLWKH 53
HST Q S+I+LW H
Sbjct: 245 HSTDQLSKSSISLWLH 198
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.128 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,193,334
Number of Sequences: 28460
Number of extensions: 43468
Number of successful extensions: 215
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 213
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 215
length of query: 173
length of database: 4,897,600
effective HSP length: 84
effective length of query: 89
effective length of database: 2,506,960
effective search space: 223119440
effective search space used: 223119440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0042.2


