

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0042.15
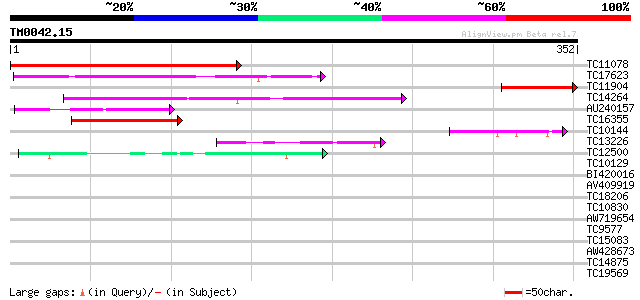
(352 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11078 similar to UP|Q40451 (Q40451) DNA-binding protein, parti... 299 5e-82
TC17623 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, part... 132 7e-32
TC11904 102 7e-23
TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), par... 96 7e-21
AU240157 77 6e-15
TC16355 similar to UP|Q9M4U6 (Q9M4U6) Histone H1 variant, partia... 60 6e-10
TC10144 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, part... 57 5e-09
TC13226 similar to UP|HMGB_SOYBN (Q10370) HMG-Y related protein ... 42 2e-04
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 41 3e-04
TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%) 38 0.002
BI420016 35 0.015
AV409919 35 0.019
TC18206 35 0.025
TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, ... 35 0.025
AW719654 34 0.042
TC9577 weakly similar to UP|ATPD_PEA (Q02758) ATP synthase delta... 34 0.042
TC15083 similar to UP|GAR1_YEAST (P28007) snoRNP protein GAR1, p... 34 0.042
AW428673 33 0.072
TC14875 similar to UP|Q9VNX6 (Q9VNX6) CG7421-PA, partial (4%) 33 0.072
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 28 3.0
>TC11078 similar to UP|Q40451 (Q40451) DNA-binding protein, partial (16%)
Length = 507
Score = 299 bits (765), Expect = 5e-82
Identities = 144/144 (100%), Positives = 144/144 (100%)
Frame = +3
Query: 1 MDSIWVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDG 60
MDSIWVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDG
Sbjct: 75 MDSIWVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDG 254
Query: 61 SSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAA 120
SSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAA
Sbjct: 255 SSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAA 434
Query: 121 QPVRRGRPPRSGIPKRRGRPPKPK 144
QPVRRGRPPRSGIPKRRGRPPKPK
Sbjct: 435 QPVRRGRPPRSGIPKRRGRPPKPK 506
>TC17623 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, partial (19%)
Length = 624
Score = 132 bits (333), Expect = 7e-32
Identities = 89/198 (44%), Positives = 104/198 (51%), Gaps = 4/198 (2%)
Frame = +1
Query: 3 SIWVPPPPPPPTA-VPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGS 61
SI P PPTA VPF + NH P P ++ A HPPYAEMIY AI ALKEKDGS
Sbjct: 25 SISSPSASAPPTATVPFAADPNNHPPPPPAPADNPA---HPPYAEMIYTAIGALKEKDGS 195
Query: 62 SKTAIAKYIEEAYTD-LPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAA 120
SK AI KYIE+ Y D L +H +LLTHHLKRLK G+L+M KKSYKLP S P +
Sbjct: 196 SKRAINKYIEQVYKDQLTQSHESLLTHHLKRLKTNGMLVMDKKSYKLPGSAPPILP---- 363
Query: 121 QPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRR--PPKHQLQATVIPFADPSLAQPPVPV 178
PP I G P P S G R+ PP+H Q + A L P
Sbjct: 364 -------PPPENIAAGAGAAPSPASRPRGRPRKVQPPQHLPQ--TLTLAGVQLQPQLPPQ 516
Query: 179 GSPRPRGRPRKNAAALPP 196
+P +P+ LPP
Sbjct: 517 QQVQPEAQPQ--TQNLPP 564
>TC11904
Length = 504
Score = 102 bits (255), Expect = 7e-23
Identities = 47/47 (100%), Positives = 47/47 (100%)
Frame = +3
Query: 306 KIPEQLPLQEHLLPQPLPEPQITYQQQHLQTQLHQLFQFQPHASFPS 352
KIPEQLPLQEHLLPQPLPEPQITYQQQHLQTQLHQLFQFQPHASFPS
Sbjct: 3 KIPEQLPLQEHLLPQPLPEPQITYQQQHLQTQLHQLFQFQPHASFPS 143
>TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), partial (76%)
Length = 1277
Score = 96.3 bits (238), Expect = 7e-21
Identities = 68/218 (31%), Positives = 102/218 (46%), Gaps = 5/218 (2%)
Frame = +2
Query: 34 NSSAPANHPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLK 93
+++ A+HP Y EM+ AI LKEK GSS+ AIAK++EE + LP LL HLK+L
Sbjct: 218 SAAKKASHPSYEEMVKDAIVTLKEKSGSSQYAIAKFLEEKHKQLPSNFKKLLLFHLKKLV 397
Query: 94 DTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRGRPPRSGIPKRRGRP---PKPKSLSNGL 150
G L+ +K S+KLPS+ S A++P + P+ K + +P KP + +
Sbjct: 398 AAGKLVKVKNSFKLPSA-KSSAAAAASKPAAAKKKPQPAAAKSKPKPKAKAKPAAAATKA 574
Query: 151 KRRP-PKHQLQATVIPFADPSLAQPPVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMV 209
K +P PK + AT P + +P+ +P A AV +
Sbjct: 575 KAKPAPKSKAAAT-------KTTAKAKPAAAAKPKAKPAAKAKPAAKAKAVAAPAKAKAS 733
Query: 210 PGRPQ-KLAVRGRPKNPAGRPVGRPKGTTSANIAQRIA 246
P +P+ K + P+ PV + K A A R A
Sbjct: 734 PAKPKAKAKSKTAPRMNVNPPVPKAKPAAKAKPAARPA 847
>AU240157
Length = 300
Score = 76.6 bits (187), Expect = 6e-15
Identities = 39/99 (39%), Positives = 58/99 (58%)
Frame = +2
Query: 4 IWVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSK 63
+ P P P+A E +N P++ PPY EMI +AIEA+ + G++K
Sbjct: 41 VQAPEPAASPSATSMATEELNK------------PSSLPPYPEMIMKAIEAVGDT-GANK 181
Query: 64 TAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLK 102
+AI+ YIE Y D+P H+TLLTHHL ++KD+G ++ K
Sbjct: 182 SAISNYIESTYGDVPAGHTTLLTHHLNQMKDSGEIVFFK 298
>TC16355 similar to UP|Q9M4U6 (Q9M4U6) Histone H1 variant, partial (50%)
Length = 584
Score = 60.1 bits (144), Expect = 6e-10
Identities = 34/70 (48%), Positives = 44/70 (62%), Gaps = 1/70 (1%)
Frame = +1
Query: 39 ANHPPYAEMIYRAIEALKEKDGSSKTAIAKYIEEAYTDLPPAH-STLLTHHLKRLKDTGL 97
A+HPPY +MI A+ AL EK GSS AIAKY+EE + + PA+ +L LK G
Sbjct: 202 ASHPPYFQMIEEALLALNEKGGSSPYAIAKYMEEKHKSVLPANFKKILGLQLKNQAAKGK 381
Query: 98 LIMLKKSYKL 107
L+ +K SYKL
Sbjct: 382 LVKIKASYKL 411
>TC10144 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, partial (8%)
Length = 600
Score = 57.0 bits (136), Expect = 5e-09
Identities = 42/89 (47%), Positives = 50/89 (55%), Gaps = 16/89 (17%)
Frame = +3
Query: 274 HASPVPAIAAIQELEIVATSDLKAPLRD--VPSQKIPEQLPL---------QEHLLP-QP 321
H SPV AIAAIQELE + DL APLRD +P Q+ P+ LP Q+ + P Q
Sbjct: 3 HESPVTAIAAIQELEELGNMDLNAPLRDETLPQQQQPQVLPQEAQPQQQVPQQQVPPLQQ 182
Query: 322 LPEPQITYQQQ----HLQTQLHQLFQFQP 346
+P Q YQQQ H+Q Q H L Q QP
Sbjct: 183 VPPQQPAYQQQYPPFHMQ-QYHHLQQQQP 266
>TC13226 similar to UP|HMGB_SOYBN (Q10370) HMG-Y related protein B (SB16B
protein) (Fragment), partial (60%)
Length = 474
Score = 42.0 bits (97), Expect = 2e-04
Identities = 36/110 (32%), Positives = 45/110 (40%), Gaps = 5/110 (4%)
Frame = +3
Query: 129 PRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGRPR 188
P + + RG PPKPK+ L V P PRPRGRP
Sbjct: 6 PNAPXKRGRGXPPKPKAAL----------PLDTVVAP---------------PRPRGRPP 110
Query: 189 KNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNP-----AGRPVGRP 233
K+ A P P V +S GRP+K+A +P +GRP GRP
Sbjct: 111 KDPNAEPKPKVVS--IASGRPRGRPKKVARSAAAASPGAAVSSGRPRGRP 254
Score = 38.9 bits (89), Expect = 0.001
Identities = 32/86 (37%), Positives = 40/86 (46%), Gaps = 2/86 (2%)
Frame = +3
Query: 106 KLPSSLPSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPK--SLSNGLKRRPPKHQLQATV 163
K ++LP D A P RGRPP K PKPK S+++G R PK ++
Sbjct: 45 KPKAALPLDTV--VAPPRPRGRPP-----KDPNAEPKPKVVSIASGRPRGRPKKVARSA- 200
Query: 164 IPFADPSLAQPPVPVGSPRPRGRPRK 189
+ A P V S RPRGRP K
Sbjct: 201 ------AAASPGAAVSSGRPRGRPPK 260
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 41.2 bits (95), Expect = 3e-04
Identities = 48/200 (24%), Positives = 66/200 (33%), Gaps = 8/200 (4%)
Frame = +1
Query: 6 VPPPPPPPTAVPFTQEAI-----NHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDG 60
+PPPPPPP+A AI D ++P + S P PP +
Sbjct: 82 LPPPPPPPSAPFIAGSAIISKVSKSVGDVSLPCSPSPPPPQPPPPSLP------------ 225
Query: 61 SSKTAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAA 120
PP S + G IM ++ P SLPS
Sbjct: 226 --------------PTTPPPASVI----------AGSAIMSGEA-SFPCSLPSP------ 312
Query: 121 QPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPS---LAQPPVP 177
P PP+ G+ PP S ++ PP L + P S + Q P+P
Sbjct: 313 -PQPPPPPPKYGVTTIPPSPPLLMSATDRAPLPPPSTHLTSLTTPSPPSSRTTITQTPLP 489
Query: 178 VGSPRPRGRPRKNAAALPPP 197
P P+ A +PPP
Sbjct: 490 SPPQPPSPPPKYGVATIPPP 549
>TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%)
Length = 562
Score = 38.1 bits (87), Expect = 0.002
Identities = 47/189 (24%), Positives = 66/189 (34%), Gaps = 13/189 (6%)
Frame = +1
Query: 6 VPPPP--------PPPTAVPFTQEAINHAPDSNIPINSSAPA--NHPPYAEMIYRAIEAL 55
+P PP PP T +P + +++P N+P AP+ NHPPY +
Sbjct: 28 IPTPPKTPSPGNQPPHTPIPPKTPSPSYSPP-NVPSPPKAPSPNNHPPYTPTPPKT---- 192
Query: 56 KEKDGSSKTAIAKYIEEAYTDLPPAHSTLLTH--HLKRLKDTGLLIMLKKSYKLPSSLPS 113
S T+ + Y LPP + ++ H+ T I P PS
Sbjct: 193 -----PSPTS-----QPPYIPLPPKTPSPISQPPHVPTPPSTPSPISQPPYTPTPPKTPS 342
Query: 114 DITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRP-PKHQLQATVIPFADPSLA 172
+Q P P P PPK S +N P P + T P PS
Sbjct: 343 PTSQPPHTPTPPNTPSPISQPPYTPTPPKTPSPTNQPPHIPSPPNSSSPTSQPPNTPSPP 522
Query: 173 QPPVPVGSP 181
+ P P P
Sbjct: 523 KTPSPTSQP 549
Score = 34.3 bits (77), Expect = 0.032
Identities = 56/234 (23%), Positives = 74/234 (30%), Gaps = 1/234 (0%)
Frame = +1
Query: 4 IWVPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSK 63
++ PP P P P T N P + IP + +P+ PP S
Sbjct: 10 VYTPPSIPTP---PKTPSPGNQPPHTPIPPKTPSPSYSPPNVP--------------SPP 138
Query: 64 TAIAKYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPV 123
A + YT PP + + LP PS I+Q P
Sbjct: 139 KAPSPNNHPPYTPTPPKTPSPTSQ--------------PPYIPLPPKTPSPISQPPHVPT 276
Query: 124 RRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPS-LAQPPVPVGSPR 182
P P PPK S P Q T P PS ++QPP +P
Sbjct: 277 PPSTPSPISQPPYTPTPPKTPS---------PTSQPPHTPTPPNTPSPISQPPY---TPT 420
Query: 183 PRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRGRPKNPAGRPVGRPKGT 236
P P+ + PPH +SS P PK P+ P +P T
Sbjct: 421 P---PKTPSPTNQPPHIPSPPNSSSPTSQPPN---TPSPPKTPS--PTSQPPNT 558
>BI420016
Length = 559
Score = 35.4 bits (80), Expect = 0.015
Identities = 28/104 (26%), Positives = 36/104 (33%), Gaps = 5/104 (4%)
Frame = +2
Query: 112 PSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSL 171
P + + Q G P +P PP +L+ PP A + P + PS
Sbjct: 161 PQTLKKPTTQCTHGGNPSPKPVPASTPSPPSSPTLTP--PPSPPSPTPNAPLSPSSPPSP 334
Query: 172 AQPPVPVGSPRPRGRPRKNAAALP-----PPHAVGGVDSSLMVP 210
PP P SP P P + P P A SSL P
Sbjct: 335 PTPPSPPPSPAPNPTPSATGSTTPSSPLTPTSASLSSPSSLFSP 466
>AV409919
Length = 423
Score = 35.0 bits (79), Expect = 0.019
Identities = 33/98 (33%), Positives = 37/98 (37%), Gaps = 3/98 (3%)
Frame = +3
Query: 105 YKLPSSLPSDITQNAAQPVRRGRPPRS-GIPKRRGRPPKPKSLSNGLKRRPPKHQLQATV 163
Y PSS PS + + A R G PP S P R P P S PP A
Sbjct: 177 YFSPSSSPSSTSSSTAGARRFGTPPLSTSSPPPRPPPSSPSS--------PPPSTSSA-- 326
Query: 164 IPFADPSLAQPPVPVGSPRPRGRPRKNAAAL--PPPHA 199
S + PP SP R PRK L PPH+
Sbjct: 327 ------SSSAPPPTTTSPTTRSWPRKIPVTLDHAPPHS 422
Score = 26.2 bits (56), Expect = 8.8
Identities = 14/37 (37%), Positives = 18/37 (47%)
Frame = +3
Query: 7 PPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPP 43
PPP PPP+ +P S P SSA ++ PP
Sbjct: 267 PPPRPPPS-----------SPSSPPPSTSSASSSAPP 344
>TC18206
Length = 709
Score = 34.7 bits (78), Expect = 0.025
Identities = 38/130 (29%), Positives = 53/130 (40%)
Frame = -1
Query: 68 KYIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRGR 127
K IE Y +T H+KR + +LI K YKLP ++ + + +RR
Sbjct: 448 KEIENVYKSSKQLEKRKITLHVKRSTKSTVLIDSSKIYKLPGTVRAKRRRCREVRLRR-- 275
Query: 128 PPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGRP 187
R G RR R + SL R +Q T + PP P G+PR R R
Sbjct: 274 --RGGPNIRRRRNLRWHSLIVPPPVRASPNQRTQTKSNIQTRHVHPPPYP-GAPR-RRRH 107
Query: 188 RKNAAALPPP 197
++ PPP
Sbjct: 106 HQSRRRPPPP 77
>TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, partial
(24%)
Length = 702
Score = 34.7 bits (78), Expect = 0.025
Identities = 37/128 (28%), Positives = 46/128 (35%), Gaps = 7/128 (5%)
Frame = +1
Query: 77 LPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRGRPPRSGIP-- 134
LPP HST L H R T LL L + ++ RR PP+ +P
Sbjct: 163 LPPPHSTSLHSHQPRFPPTNLL--QSHFRHLQQTHHMELHHPPLHLARRHLPPKPRLPPR 336
Query: 135 KRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRG-----RPRK 189
R+ RPP R PP P P A P P P PR +P +
Sbjct: 337 PRKSRPP----------RLPP---------PLNLPHSAPCPQPQKQPLPRPHTKPLQPHR 459
Query: 190 NAAALPPP 197
+ LP P
Sbjct: 460 HETPLPLP 483
>AW719654
Length = 491
Score = 33.9 bits (76), Expect = 0.042
Identities = 26/89 (29%), Positives = 32/89 (35%), Gaps = 3/89 (3%)
Frame = +2
Query: 112 PSDITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVI---PFAD 168
P +T + R R P +P PP P S S PP AT P +
Sbjct: 128 PLLLTSPPSLSYTRSRHPPPWLP*HSPPPPSPPSASATATATPPNRHSSATPSN*NPPSL 307
Query: 169 PSLAQPPVPVGSPRPRGRPRKNAAALPPP 197
P L + P P P P + LPPP
Sbjct: 308 PFLQRSHSPPSLPFPENTPLSLSEPLPPP 394
>TC9577 weakly similar to UP|ATPD_PEA (Q02758) ATP synthase delta chain,
chloroplast precursor , partial (54%)
Length = 627
Score = 33.9 bits (76), Expect = 0.042
Identities = 25/84 (29%), Positives = 30/84 (34%)
Frame = +3
Query: 127 RPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQPPVPVGSPRPRGR 186
R P S P R PP P S SN + PP A ++P PP P P
Sbjct: 141 RNPSSTSPSRA--PPSPHSTSNLSQSPPPA----APAAAHSEPE*TPPPPPATPPHSPTS 302
Query: 187 PRKNAAALPPPHAVGGVDSSLMVP 210
P + PPP S + P
Sbjct: 303 PAPTNPSTPPPPTWRSSRRSFLTP 374
>TC15083 similar to UP|GAR1_YEAST (P28007) snoRNP protein GAR1, partial (7%)
Length = 1367
Score = 33.9 bits (76), Expect = 0.042
Identities = 30/114 (26%), Positives = 45/114 (39%), Gaps = 4/114 (3%)
Frame = -2
Query: 6 VPPPPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTA 65
VPP P PT P P N P SAP+NHP A D +
Sbjct: 1270 VPPTPAGPTPFP---------PP*NSPSRPSAPSNHPTPCPPSRHAAAQNAPADAAVNLP 1118
Query: 66 IAKYIEEAYT-DLPPAHSTLL---THHLKRLKDTGLLIMLKKSYKLPSSLPSDI 115
++ +A + +LPP+ LL +HH L+ + + + P+ PS +
Sbjct: 1117 VSPPSNKAVSPNLPPSPPCLLFPSSHH*FHLQYSSARVGIYPDSPAPTPTPSTL 956
>AW428673
Length = 352
Score = 33.1 bits (74), Expect = 0.072
Identities = 26/90 (28%), Positives = 35/90 (38%), Gaps = 14/90 (15%)
Frame = +1
Query: 122 PVRRGRPPRSGIPKRR---GRPPKPKSLSNGLKRRPPKHQLQATVIPFA----------- 167
PVRR P P +R PP+ + ++R PPK +L + P
Sbjct: 61 PVRRCPPNDRKCPPKRELHSPPPRVRRCLPNVRRCPPKPELHSPPPPVRRCPPNDRRCPP 240
Query: 168 DPSLAQPPVPVGSPRPRGRPRKNAAALPPP 197
P L PP PV + P P + PPP
Sbjct: 241 KPVLHSPPPPVHTSPPNDIPHPIHHSPPPP 330
>TC14875 similar to UP|Q9VNX6 (Q9VNX6) CG7421-PA, partial (4%)
Length = 1240
Score = 33.1 bits (74), Expect = 0.072
Identities = 42/134 (31%), Positives = 53/134 (39%), Gaps = 2/134 (1%)
Frame = +2
Query: 115 ITQNAAQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPFADPSLAQP 174
I + QP R R P P RR +PP P + +R H LQ + P Q
Sbjct: 146 IQRRQHQPRHRRRRPPLPPP*RRQQPPPPHL--HPRRRLLHLHPLQQRL-----PPPPQQ 304
Query: 175 PVPVGSPRPRGRPRKNAAALPPPHAVGGVDSSLMVPGRPQKLAVRG--RPKNPAGRPVGR 232
G R R RP PPPH + S V RP + +G R ++ RPV
Sbjct: 305 RRREGKRRRRLRPLPPRPGAPPPHRLRRHLGSHPVGCRPPRPVAQGPRRSRSCFLRPVPP 484
Query: 233 PKGTTSANIAQRIA 246
G ANIA +A
Sbjct: 485 RAG---ANIAHSMA 517
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 27.7 bits (60), Expect = 3.0
Identities = 22/101 (21%), Positives = 35/101 (33%), Gaps = 11/101 (10%)
Frame = +1
Query: 108 PSSLPSDITQNA-AQPVRRGRPPRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIPF 166
P P+ +T+N P + P S + P P + S ++P P
Sbjct: 139 PPPSPTKVTENPPTAPSTSTKSPTSSASEE----PSPPNPSTSTTQQPKHSPTSPPASPS 306
Query: 167 ADPSLAQPPVPVGSP----------RPRGRPRKNAAALPPP 197
P +PP + SP P +A++ PPP
Sbjct: 307 PSPPSTKPPTAMASPSTSPLSATKSHPTPPAAPSASSTPPP 429
Score = 26.6 bits (57), Expect = 6.7
Identities = 33/151 (21%), Positives = 59/151 (38%), Gaps = 17/151 (11%)
Frame = +2
Query: 6 VPPPPPPPTAVPFTQEAINHAPDSNIPI--NSSAPANHPPYAEMIYRAIEALKEKDGSSK 63
+PPPPPPPT +H P++ + + + H + R I + S
Sbjct: 44 LPPPPPPPT------HHHHHLPNNPLTLLQHHQLRRRHHRHRLRR*RKIHQRLHRPQQSL 205
Query: 64 TAIAKYIEEAYTDLPP----------AHSTLLTHHLKRLKD--TGLLIMLKKSYKLPSSL 111
+ + PP H L HHL+ ++ L +L + +LP+
Sbjct: 206 LLLPRRKSHLRPTPPPLRPNNQNTHRLHHPLHLHHLRHQQNLLRRWLRLLPRPSRLPN-- 379
Query: 112 PSDITQNAAQPVRR---GRPPRSGIPKRRGR 139
P+ + + +P++R +PP +RR R
Sbjct: 380 PTQLRRRHLRPLQRHHQQQPPSKPCRRRRVR 472
Score = 26.2 bits (56), Expect = 8.8
Identities = 33/157 (21%), Positives = 49/157 (31%)
Frame = +1
Query: 9 PPPPPTAVPFTQEAINHAPDSNIPINSSAPANHPPYAEMIYRAIEALKEKDGSSKTAIAK 68
PPPPP+ P + + + P + + +PP A S+K+ +
Sbjct: 73 PPPPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTA------------PSTSTKSPTSS 216
Query: 69 YIEEAYTDLPPAHSTLLTHHLKRLKDTGLLIMLKKSYKLPSSLPSDITQNAAQPVRRGRP 128
EE PP ST T K S P + PS + P P
Sbjct: 217 ASEE---PSPPNPSTSTTQQPKH------------SPTSPPASPSPSPPSTKPPTAMASP 351
Query: 129 PRSGIPKRRGRPPKPKSLSNGLKRRPPKHQLQATVIP 165
S + + P P + S P L+ P
Sbjct: 352 STSPLSATKSHPTPPAAPSASSTPPPTTTSLKTMSSP 462
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,194,333
Number of Sequences: 28460
Number of extensions: 150994
Number of successful extensions: 4132
Number of sequences better than 10.0: 339
Number of HSP's better than 10.0 without gapping: 2193
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3265
length of query: 352
length of database: 4,897,600
effective HSP length: 91
effective length of query: 261
effective length of database: 2,307,740
effective search space: 602320140
effective search space used: 602320140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0042.15


