

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
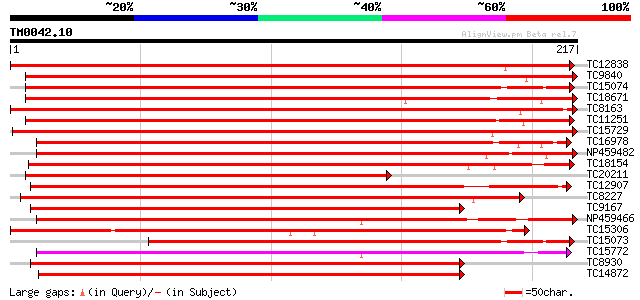
Query= TM0042.10
(217 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12838 UP|Q40196 (Q40196) RAB11F, complete 340 1e-94
TC9840 UP|R11D_LOTJA (Q40194) Ras-related protein Rab11D, complete 231 6e-62
TC15074 similar to UP|R11B_TOBAC (Q40521) Ras-related protein Ra... 227 1e-60
TC18671 UP|Q40198 (Q40198) RAB11H, complete 226 2e-60
TC8163 UP|R11C_LOTJA (Q40193) Ras-related protein Rab11C, complete 224 9e-60
TC11251 UP|R11E_LOTJA (Q40195) Ras-related protein Rab11E, complete 223 3e-59
TC15729 homologue to UP|RB1C_ARATH (O04486) Ras-related protein ... 221 6e-59
TC16978 UP|R11A_LOTJA (Q40191) Ras-related protein Rab11A, complete 207 1e-54
NP459482 RAB11J [Lotus japonicus] 203 2e-53
TC18154 UP|Q40192 (Q40192) RAB11B, complete 199 3e-52
TC20211 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, p... 187 2e-48
TC12907 UP|Q40201 (Q40201) RAB1A, complete 173 2e-44
TC8227 UP|Q40203 (Q40203) RAB1C, complete 172 4e-44
TC9167 UP|Q40202 (Q40202) RAB1B (Fragment), complete 170 2e-43
NP459466 RAB8C [Lotus japonicus] 168 6e-43
TC15306 UP|Q40207 (Q40207) RAB1Y (Fragment), complete 165 6e-42
TC15073 UP|Q40199 (Q40199) RAB11I (Fragment), complete 165 6e-42
TC15772 UP|Q40218 (Q40218) RAB8D, complete 164 8e-42
TC8930 UP|Q40204 (Q40204) RAB1D, complete 164 8e-42
TC14872 UP|Q40208 (Q40208) RAB2A, complete 164 8e-42
>TC12838 UP|Q40196 (Q40196) RAB11F, complete
Length = 1099
Score = 340 bits (871), Expect = 1e-94
Identities = 175/217 (80%), Positives = 189/217 (86%), Gaps = 1/217 (0%)
Frame = +2
Query: 1 MMADAFDEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDK 60
M ADAFDE+CDYLFK VLIGDSGVGKSNL+SRFAKDEFRLDSKPTIGVEFAY NIKVRDK
Sbjct: 110 MAADAFDEQCDYLFKAVLIGDSGVGKSNLISRFAKDEFRLDSKPTIGVEFAYRNIKVRDK 289
Query: 61 LIKAQIWDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMV 120
LIKAQIWDTAGQERFRAITSSYYRGALG M+VYDITRR ++ NV WL ELREFG EDMV
Sbjct: 290 LIKAQIWDTAGQERFRAITSSYYRGALGPMLVYDITRRPTFMNVIKWLHELREFGNEDMV 469
Query: 121 VILVGNKSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQ 180
V+LV NKSDL QSREVEKEEG+ FAE EGLCFMETSAL+NLNVEE FLEMITKI DI Q
Sbjct: 470 VVLVANKSDLVQSREVEKEEGEEFAEIEGLCFMETSALQNLNVEEAFLEMITKIHDIISQ 649
Query: 181 RSLEAKVD-EKPINVFSGKEIHLVDEVTATKQATCCS 216
+SLE K++ ++V GKEI + DEVTATKQA CCS
Sbjct: 650 KSLETKMNGPTTLSVPIGKEIQVADEVTATKQANCCS 760
>TC9840 UP|R11D_LOTJA (Q40194) Ras-related protein Rab11D, complete
Length = 1302
Score = 231 bits (590), Expect = 6e-62
Identities = 120/212 (56%), Positives = 156/212 (72%), Gaps = 1/212 (0%)
Frame = +2
Query: 7 DEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQI 66
D+E DYLFK VLIGDSGVGKSNLLSRF K+EF L+SK TIGVEFA + V K++KAQI
Sbjct: 266 DDEYDYLFKLVLIGDSGVGKSNLLSRFTKNEFNLESKSTIGVEFATKTLNVDAKVVKAQI 445
Query: 67 WDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGN 126
WDTAGQER+RAITS+YYRGA+GA++VYD+TRR+++EN WL ELR+ ++VV+L+GN
Sbjct: 446 WDTAGQERYRAITSAYYRGAVGALLVYDVTRRATFENAARWLKELRDHTDPNIVVMLIGN 625
Query: 127 KSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQRSLEAK 186
KSDL V E+GK FAE E L FMETSAL+ NVE F E++T+I I +R++EA
Sbjct: 626 KSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLTQIYRIVSKRAVEAG 805
Query: 187 VDEKPINVFS-GKEIHLVDEVTATKQATCCST 217
+ S G+ I++ ++ + K+ CCST
Sbjct: 806 DSGSSSGLPSKGQTINVKEDSSVLKRFGCCST 901
>TC15074 similar to UP|R11B_TOBAC (Q40521) Ras-related protein Rab11B,
partial (89%)
Length = 973
Score = 227 bits (578), Expect = 1e-60
Identities = 115/210 (54%), Positives = 157/210 (74%)
Frame = +3
Query: 7 DEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQI 66
DE+ DYLFK VLIGDSGVGKSNLLSRF ++EF L+SK TIGVEFA ++++ DK +KAQI
Sbjct: 180 DEDYDYLFKLVLIGDSGVGKSNLLSRFTRNEFNLESKSTIGVEFATRSVRIHDKTVKAQI 359
Query: 67 WDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGN 126
WDTAGQER+RAITS+YYRGA+GA++VYD+TR+ ++ENV+ WL ELR+ ++V++LVGN
Sbjct: 360 WDTAGQERYRAITSAYYRGAVGALIVYDVTRQVTFENVQRWLKELRDHTDANIVIMLVGN 539
Query: 127 KSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQRSLEAK 186
K+DL R V EE FAE E FMETSAL++LNV+ F+ ++++I ++ +++L
Sbjct: 540 KADLRHLRAVPTEEATAFAERENTYFMETSALESLNVDNAFIXVLSEIYNVVSRKTLXKG 719
Query: 187 VDEKPINVFSGKEIHLVDEVTATKQATCCS 216
D P + G+ I+L D V+ K CCS
Sbjct: 720 ND--PGALPQGQTINLGD-VSXXKXPGCCS 800
>TC18671 UP|Q40198 (Q40198) RAB11H, complete
Length = 1181
Score = 226 bits (577), Expect = 2e-60
Identities = 118/214 (55%), Positives = 160/214 (74%), Gaps = 3/214 (1%)
Frame = +3
Query: 7 DEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQI 66
D++ DYLFK VLIGDSGVGKSNLLSRF K+EF L+SK TIGVEFA +I+V DK++KAQI
Sbjct: 222 DDDYDYLFKVVLIGDSGVGKSNLLSRFTKNEFSLESKSTIGVEFATRSIRVDDKVVKAQI 401
Query: 67 WDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGN 126
WDTAGQER+RAITS+YYRGA+GA++VYD+TR ++ENV WL ELR+ ++VV+LVGN
Sbjct: 402 WDTAGQERYRAITSAYYRGAVGALLVYDVTRHVTFENVERWLKELRDHTDTNIVVMLVGN 581
Query: 127 KSDLGQSREVEKEEGKGFAETEGL-CFMETSALKNLNVEEVFLEMITKILDITCQRSLEA 185
K+DL R V E+ GF E FMETSAL++LNVE F E++T+I + +++LE
Sbjct: 582 KADLRHLRAVSTEDSSGFLLNERTHFFMETSALESLNVENAFTEVLTQIYRVVSKKALE- 758
Query: 186 KVDEKPINVFSGKEIHL--VDEVTATKQATCCST 217
+ + P + G+ I++ D+V+ K++ CCS+
Sbjct: 759 -IGDDPTALPKGQTINVGSRDDVSPVKKSGCCSS 857
>TC8163 UP|R11C_LOTJA (Q40193) Ras-related protein Rab11C, complete
Length = 1193
Score = 224 bits (571), Expect = 9e-60
Identities = 115/218 (52%), Positives = 157/218 (71%), Gaps = 1/218 (0%)
Frame = +3
Query: 1 MMADAFDEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDK 60
+MA D E DYLFK VLIGDSGVGKSN+LSRF ++EF L+SK TIGVEFA ++V K
Sbjct: 234 VMAHRVDHEYDYLFKIVLIGDSGVGKSNILSRFTRNEFCLESKSTIGVEFATRTLQVEGK 413
Query: 61 LIKAQIWDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMV 120
+KAQIWDTAGQER+RAITS+YYRGA+GA++VYDIT+R +++NV+ WL ELR+ ++V
Sbjct: 414 TVKAQIWDTAGQERYRAITSAYYRGAVGALLVYDITKRQTFDNVQRWLRELRDHADSNIV 593
Query: 121 VILVGNKSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQ 180
+++ GNKSDL R V +++G AE EGL F+ETSAL+ N+E+ F ++T+I I +
Sbjct: 594 IMMAGNKSDLNHLRAVSEDDGGALAEKEGLSFLETSALEATNIEKAFQTILTEIYHIVSK 773
Query: 181 RSLEAKVDEKPINV-FSGKEIHLVDEVTATKQATCCST 217
++L A+ +V G I++ D TK+ CCST
Sbjct: 774 KALAAQEATAGASVPGQGTTINVADTSGNTKKG-CCST 884
>TC11251 UP|R11E_LOTJA (Q40195) Ras-related protein Rab11E, complete
Length = 950
Score = 223 bits (567), Expect = 3e-59
Identities = 116/212 (54%), Positives = 154/212 (71%), Gaps = 2/212 (0%)
Frame = +2
Query: 7 DEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQI 66
D+E DYLFK VLIGDSGVGKSNLLSRF ++EF L+SK TIGVEFA ++ + K+IKAQI
Sbjct: 80 DDEYDYLFKLVLIGDSGVGKSNLLSRFTRNEFNLESKSTIGVEFATKSLNIDGKVIKAQI 259
Query: 67 WDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGN 126
WDTAGQER+RAITS+YYRGA+GA++VYD+TR +++E WL ELR+ ++VV+L+GN
Sbjct: 260 WDTAGQERYRAITSAYYRGAVGALLVYDVTRSTTFETAGRWLKELRDHTDPNIVVMLIGN 439
Query: 127 KSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQRSLEAK 186
KSDL V E+GK FAE E L FMETSAL+ NVE F E++T+I I +R++EA
Sbjct: 440 KSDLRHLVTVSTEDGKAFAEKESLYFMETSALEATNVENAFSEVLTQIYRIVSKRAVEAG 619
Query: 187 VDEKPINVF--SGKEIHLVDEVTATKQATCCS 216
D +V G+ I++ ++ + + CCS
Sbjct: 620 -DRPSTSVVPSQGQTINVNEDSSVLNRYRCCS 712
>TC15729 homologue to UP|RB1C_ARATH (O04486) Ras-related protein Rab11C,
partial (85%)
Length = 860
Score = 221 bits (564), Expect = 6e-59
Identities = 112/217 (51%), Positives = 154/217 (70%), Gaps = 1/217 (0%)
Frame = +2
Query: 2 MADAFDEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKL 61
MA +EE DYLFK VLIGDSGVGKSNLLSRF ++EF L+SK TIGVEFA ++V ++
Sbjct: 83 MARRGEEEYDYLFKVVLIGDSGVGKSNLLSRFTRNEFCLESKSTIGVEFATRTLEVEGRM 262
Query: 62 IKAQIWDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVV 121
+KAQIWDTAGQER+RAITS+YYRGALGA++VYD+T+ +S+ENV WL ELR+ ++V+
Sbjct: 263 VKAQIWDTAGQERYRAITSAYYRGALGALLVYDVTKPTSFENVTRWLKELRDHADANIVI 442
Query: 122 ILVGNKSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQR 181
+L+GNK+DL R V E+ + +AE EGL F+ETSAL+ NV++ F ++ +I I ++
Sbjct: 443 MLIGNKTDLKHLRGVATEDAQSYAEKEGLSFIETSALEATNVDKAFQTILAQIYRIISKK 622
Query: 182 SL-EAKVDEKPINVFSGKEIHLVDEVTATKQATCCST 217
SL + D N+ G I + + +CC+T
Sbjct: 623 SLSSSSTDPSAPNIKQGNTITIQGGLQPNTSKSCCTT 733
>TC16978 UP|R11A_LOTJA (Q40191) Ras-related protein Rab11A, complete
Length = 984
Score = 207 bits (527), Expect = 1e-54
Identities = 111/212 (52%), Positives = 150/212 (70%), Gaps = 7/212 (3%)
Frame = +2
Query: 11 DYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWDTA 70
DY+FK VLIGDS VGKS +L+RFA++EF LDSK TIGVEF + + K +KAQIWDTA
Sbjct: 104 DYVFKVVLIGDSAVGKSQILARFARNEFSLDSKSTIGVEFQTRTLVIDHKTVKAQIWDTA 283
Query: 71 GQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKSDL 130
GQER+RA+TS+YYRGA+GAM+VYDIT+R +++++ WL ELR +++V+IL+GNK DL
Sbjct: 284 GQERYRAVTSAYYRGAVGAMLVYDITKRQTFDHIPRWLEELRNHADKNIVIILIGNKCDL 463
Query: 131 GQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQRSLEAKVDEK 190
R+V E+ K FAE EGL F+ETSAL+ NVE F ++T+I +I ++SL A DE
Sbjct: 464 VNQRDVPTEDAKEFAEKEGLFFLETSALEATNVESAFTTVLTEIYNIVNKKSLAA--DES 637
Query: 191 PIN----VFSGKEIHL---VDEVTATKQATCC 215
N SG++I + E+ A K+ CC
Sbjct: 638 QGNGNSASLSGQKIIIPGPAQEIPA-KRNMCC 730
>NP459482 RAB11J [Lotus japonicus]
Length = 672
Score = 203 bits (516), Expect = 2e-53
Identities = 112/213 (52%), Positives = 143/213 (66%), Gaps = 6/213 (2%)
Frame = +1
Query: 11 DYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWDTA 70
DYLFK VLIGDS VGKSNLL+RFA+DEF +SK TIGVEF I + K IKAQIWDTA
Sbjct: 34 DYLFKIVLIGDSAVGKSNLLARFARDEFYPNSKSTIGVEFQTQKIDINGKEIKAQIWDTA 213
Query: 71 GQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKSDL 130
GQERFRA+TS+YYRGA+GA+VVYDI+RR +++++ WL EL ++V ILVGNKSDL
Sbjct: 214 GQERFRAVTSAYYRGAVGALVVYDISRRQTFDSIGRWLNELHTHSDMNVVTILVGNKSDL 393
Query: 131 GQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQR---SLEAKV 187
+REV EGK AE +GL FMETSAL + NV F ++ +I +I ++ S E
Sbjct: 394 KDAREVTTAEGKALAEAQGLFFMETSALDSSNVAAAFQTVVKEIYNILSRKVMMSQELTK 573
Query: 188 DEKPINVFSGKEIHLVD---EVTATKQATCCST 217
E P + +GK + + E + CCS+
Sbjct: 574 HEVP-RIENGKSVVIQGENLEADGQSKKGCCSS 669
>TC18154 UP|Q40192 (Q40192) RAB11B, complete
Length = 1183
Score = 199 bits (506), Expect = 3e-52
Identities = 108/221 (48%), Positives = 144/221 (64%), Gaps = 12/221 (5%)
Frame = +2
Query: 8 EECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIW 67
++ DY+FK V+IGDS VGK+ +LSRFAK+EF DSK TIGVEF + + K+IKAQIW
Sbjct: 353 DKIDYVFKVVVIGDSAVGKTQILSRFAKNEFCFDSKATIGVEFQTRTVTINAKVIKAQIW 532
Query: 68 DTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNK 127
DTAGQER+RA+TS+YYRGALGAM+VYDIT+R S+++V W+ ELR +V++L+GNK
Sbjct: 533 DTAGQERYRAVTSAYYRGALGAMLVYDITKRQSFDHVARWVEELRAHADSSIVIMLIGNK 712
Query: 128 SDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKI-LDITCQRSLE-- 184
DL R V E+ FAE +GL F ETSAL NVE FL+++ +I + +RSL
Sbjct: 713 GDLVDQRVVLSEDAVEFAEEQGLFFSETSALTGENVESAFLKLLQEINTRVVSKRSLSDC 892
Query: 185 ---------AKVDEKPINVFSGKEIHLVDEVTATKQATCCS 216
+ I+V SG E+ E+T K+ CS
Sbjct: 893 NHGKKTNGGNSLKGSKIDVISGPEL----EITEVKKVPSCS 1003
>TC20211 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, partial
(67%)
Length = 623
Score = 187 bits (474), Expect = 2e-48
Identities = 92/140 (65%), Positives = 112/140 (79%)
Frame = +1
Query: 7 DEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQI 66
D++ DYLFK VLIGDSGVGKSNLLSRF K+EF L+SK TIGVEFA + V K+IK+QI
Sbjct: 202 DDDYDYLFKVVLIGDSGVGKSNLLSRFTKNEFNLESKSTIGVEFATRTLNVDSKVIKSQI 381
Query: 67 WDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGN 126
WDTAGQER+RAITS+YYRGALGA++VYD+TR +++ENV WL ELR ++VV+LVGN
Sbjct: 382 WDTAGQERYRAITSAYYRGALGALLVYDVTRHATFENVDRWLKELRNHTDSNIVVMLVGN 561
Query: 127 KSDLGQSREVEKEEGKGFAE 146
KSDL V E+GK +AE
Sbjct: 562 KSDLRHLVAVSTEDGKSYAE 621
>TC12907 UP|Q40201 (Q40201) RAB1A, complete
Length = 869
Score = 173 bits (438), Expect = 2e-44
Identities = 92/207 (44%), Positives = 129/207 (61%)
Frame = +2
Query: 9 ECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWD 68
E DYLFK +LIGDS VGKS LL RFA D + TIGV+F ++++ K IK QIWD
Sbjct: 122 EYDYLFKLLLIGDSSVGKSCLLLRFADDSYVDSYISTIGVDFKIRTVELQGKTIKLQIWD 301
Query: 69 TAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKS 128
TAGQERFR ITSSYYRGA G ++VYD+T S+ NV+ WL E+ + + + +LVGNKS
Sbjct: 302 TAGQERFRTITSSYYRGAHGIIIVYDVTEMESFNNVKQWLNEIDRYANDSVCKLLVGNKS 481
Query: 129 DLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQRSLEAKVD 188
DL +++ V+ + K FA+ G+ F+ETSA ++NVE+ FL M +I + K+
Sbjct: 482 DLVENKVVDTQTAKAFADELGIPFLETSAKDSINVEQAFLTMAAEI---------KKKMG 634
Query: 189 EKPINVFSGKEIHLVDEVTATKQATCC 215
+P S + + + + K + CC
Sbjct: 635 SQPTGSKSAETVQMKGQPIPQK-SNCC 712
>TC8227 UP|Q40203 (Q40203) RAB1C, complete
Length = 1083
Score = 172 bits (436), Expect = 4e-44
Identities = 95/199 (47%), Positives = 124/199 (61%), Gaps = 6/199 (3%)
Frame = +2
Query: 5 AFDEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKA 64
A + E DYLFK +LIGDSGVGKS LL RFA D + TIGV+F ++ K IK
Sbjct: 125 AMNPEYDYLFKLLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKL 304
Query: 65 QIWDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILV 124
QIWDTAGQERFR ITSSYYRGA G +VVYD+T + S+ NV+ WL E+ + E++ +LV
Sbjct: 305 QIWDTAGQERFRTITSSYYRGAHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLV 484
Query: 125 GNKSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILD------IT 178
GNK DL ++ V E K FA+ G+ FMETSA NVE+ F+ M +I + +
Sbjct: 485 GNKCDLTANKVVSYETAKAFADEIGIPFMETSAKNATNVEQAFMAMAAEIKNRMASQPVN 664
Query: 179 CQRSLEAKVDEKPINVFSG 197
R ++ +P+N SG
Sbjct: 665 NARPPTVQIRGQPVNQKSG 721
>TC9167 UP|Q40202 (Q40202) RAB1B (Fragment), complete
Length = 951
Score = 170 bits (431), Expect = 2e-43
Identities = 89/166 (53%), Positives = 110/166 (65%)
Frame = +2
Query: 9 ECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWD 68
E DYLFK +LIGDSGVGKS LL RFA D + TIGV+F ++ K IK QIWD
Sbjct: 101 EYDYLFKLLLIGDSGVGKSCLLLRFADDSYIESYISTIGVDFKIRTVEQDAKTIKLQIWD 280
Query: 69 TAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKS 128
TAGQERFR ITSSYYRGA G ++VYD+T S+ NV+ WL E+ + +++ +LVGNKS
Sbjct: 281 TAGQERFRTITSSYYRGAHGIIIVYDVTDEESFNNVKQWLSEIDRYASDNVNKLLVGNKS 460
Query: 129 DLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKI 174
DL +R V E K FA+ G+ FMETSA NVE+ F+ M I
Sbjct: 461 DLTANRAVSYETAKAFADEIGIPFMETSAKDATNVEQAFMAMAASI 598
>NP459466 RAB8C [Lotus japonicus]
Length = 639
Score = 168 bits (426), Expect = 6e-43
Identities = 92/208 (44%), Positives = 128/208 (61%), Gaps = 1/208 (0%)
Frame = +1
Query: 11 DYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWDTA 70
DYL K +LIGDSGVGKS LL RF+ F TIG++F I++ K IK QIWDTA
Sbjct: 37 DYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTA 216
Query: 71 GQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKSDL 130
GQER R IT++YYRGA G ++VYD+T SS+ N+RNW+ + + +++ ILVGNK+D+
Sbjct: 217 GQERVRTITTAYYRGAKGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADM 396
Query: 131 GQS-REVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQRSLEAKVDE 189
+S R V +G+ A+ G+ F ETSA NLNVEEVF + I QR +
Sbjct: 397 DESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIK----QRLADTDSKA 564
Query: 190 KPINVFSGKEIHLVDEVTATKQATCCST 217
+P + +I+ D+ A + +TCC +
Sbjct: 565 EPQTI----KINQPDQPAAAQNSTCCGS 636
>TC15306 UP|Q40207 (Q40207) RAB1Y (Fragment), complete
Length = 1019
Score = 165 bits (417), Expect = 6e-42
Identities = 92/201 (45%), Positives = 127/201 (62%), Gaps = 2/201 (0%)
Frame = +2
Query: 1 MMADAFDEECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDK 60
M + + +E DYLFK ++IGDSGVGKS+LL F DEF+ D PTIGV+F + + K
Sbjct: 74 MDSSSAPQEFDYLFKLLMIGDSGVGKSSLLLSFTSDEFQ-DMSPTIGVDFKVKYVAIGGK 250
Query: 61 LIKAQIWDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRN-WLLELREFG-GED 118
+K IWDTAGQERFR +TSSYYRGA G ++VYD+TRR ++ N+ W E+ + +D
Sbjct: 251 KLKLAIWDTAGQERFRTLTSSYYRGAQGIIMVYDVTRRDTFTNLSEIWAKEIDLYSTNQD 430
Query: 119 MVVILVGNKSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDIT 178
+ +LVGNK D R V K+EG FA G F+E SA +NV++ F E++ KILD
Sbjct: 431 CIKMLVGNKVDKESDRVVSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTP 610
Query: 179 CQRSLEAKVDEKPINVFSGKE 199
+ +K +K N+F K+
Sbjct: 611 SLLAEGSKGVKK--NIFKDKQ 667
>TC15073 UP|Q40199 (Q40199) RAB11I (Fragment), complete
Length = 733
Score = 165 bits (417), Expect = 6e-42
Identities = 82/163 (50%), Positives = 121/163 (73%)
Frame = +1
Query: 54 NIKVRDKLIKAQIWDTAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELRE 113
++++ DK +KAQIWDTAGQER+RAITS+YYRGA+GA++VYD+TR+ ++ENV+ WL ELR+
Sbjct: 7 SVRIHDKTVKAQIWDTAGQERYRAITSAYYRGAVGALIVYDVTRQVTFENVQRWLKELRD 186
Query: 114 FGGEDMVVILVGNKSDLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITK 173
++V++LVGNK+DL R V EE FAE E FMETSAL++LNV+ F+E++++
Sbjct: 187 HTDANIVIMLVGNKADLRHLRAVPTEEATAFAERENTYFMETSALESLNVDNAFIEVLSE 366
Query: 174 ILDITCQRSLEAKVDEKPINVFSGKEIHLVDEVTATKQATCCS 216
I ++ +++LE D P + G+ I+L D V+A K+ CCS
Sbjct: 367 IYNVVSRKTLEKGND--PGALPQGQTINLGD-VSAVKKPGCCS 486
>TC15772 UP|Q40218 (Q40218) RAB8D, complete
Length = 1139
Score = 164 bits (416), Expect = 8e-42
Identities = 89/206 (43%), Positives = 124/206 (59%), Gaps = 1/206 (0%)
Frame = +2
Query: 11 DYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWDTA 70
DYL K +LIGDSGVGKS LL RF+ F TIG++F I++ K +K QIWDTA
Sbjct: 275 DYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRVKLQIWDTA 454
Query: 71 GQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKSDL 130
GQERFR IT++YYRGA+G ++VYD+T +S+ N+RNW+ + + +++ ILVGNK+D+
Sbjct: 455 GQERFRTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADM 634
Query: 131 GQS-REVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKILDITCQRSLEAKVDE 189
+S R V +G+ A+ G+ F ETSA NLNVEEVF + I +A+
Sbjct: 635 DESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDHKAEPTT 814
Query: 190 KPINVFSGKEIHLVDEVTATKQATCC 215
IN S A +++CC
Sbjct: 815 LKINQDSA-----AGAGEAANKSSCC 877
>TC8930 UP|Q40204 (Q40204) RAB1D, complete
Length = 943
Score = 164 bits (416), Expect = 8e-42
Identities = 86/166 (51%), Positives = 108/166 (64%)
Frame = +3
Query: 9 ECDYLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWD 68
E DYLFK +LIGDSGVGKS LL RF D + TIGV+F ++ K IK QIWD
Sbjct: 99 EYDYLFKLLLIGDSGVGKSCLLLRFGDDSYIESYISTIGVDFKIRTVEQDAKTIKLQIWD 278
Query: 69 TAGQERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKS 128
TAGQERFR ITSSYYRGA G ++VYD+T S+ NV+ WL E+ + +++ +LVGNK
Sbjct: 279 TAGQERFRTITSSYYRGAHGIIIVYDVTDEESFNNVKQWLSEIDRYASDNVNKLLVGNKC 458
Query: 129 DLGQSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKI 174
DL +R V + K FA+ G+ FMETSA NVE+ F+ M I
Sbjct: 459 DLTANRAVSYDTAKEFADPIGIPFMETSAKDATNVEQAFMAMSASI 596
>TC14872 UP|Q40208 (Q40208) RAB2A, complete
Length = 1125
Score = 164 bits (416), Expect = 8e-42
Identities = 84/163 (51%), Positives = 109/163 (66%)
Frame = +3
Query: 12 YLFKTVLIGDSGVGKSNLLSRFAKDEFRLDSKPTIGVEFAYSNIKVRDKLIKAQIWDTAG 71
YLFK ++IGD+GVGKS LL +F F+ TIGVEF I + +K IK QIWDTAG
Sbjct: 219 YLFKYIIIGDTGVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDNKPIKLQIWDTAG 398
Query: 72 QERFRAITSSYYRGALGAMVVYDITRRSSYENVRNWLLELREFGGEDMVVILVGNKSDLG 131
QE FR+IT SYYRGA GA++VYDITRR ++ ++ +WL + R+ +M ++L+GNK DL
Sbjct: 399 QESFRSITRSYYRGAAGALLVYDITRRETFNHLASWLEDARQHANANMTIMLIGNKCDLA 578
Query: 132 QSREVEKEEGKGFAETEGLCFMETSALKNLNVEEVFLEMITKI 174
R V EEG+ FA+ GL FME SA NVEE F++ I
Sbjct: 579 HRRAVSTEEGEQFAKEHGLIFMEASAKTAQNVEEAFIKTAATI 707
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,961,645
Number of Sequences: 28460
Number of extensions: 28399
Number of successful extensions: 212
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 188
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 188
length of query: 217
length of database: 4,897,600
effective HSP length: 87
effective length of query: 130
effective length of database: 2,421,580
effective search space: 314805400
effective search space used: 314805400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0042.10


