

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0042.1
(635 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
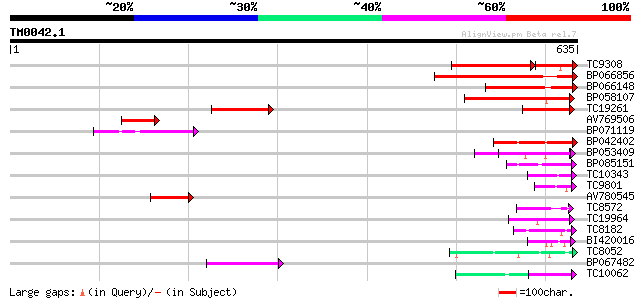
Score E
Sequences producing significant alignments: (bits) Value
TC9308 weakly similar to UP|Q9SQ55 (Q9SQ55) Nuclear RNA binding ... 180 6e-67
BP066856 233 5e-62
BP066148 184 4e-47
BP058107 179 2e-45
TC19261 similar to UP|BAD06577 (BAD06577) Cell wall protein Awa1... 93 1e-19
AV769506 91 7e-19
BP071119 85 4e-17
BP042402 79 2e-15
BP053409 68 5e-12
BP085151 64 6e-11
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 50 8e-07
TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like... 46 2e-05
AV780545 45 4e-05
TC8572 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 ho... 45 5e-05
TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%) 44 8e-05
TC8182 weakly similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, ... 43 1e-04
BI420016 43 1e-04
TC8052 similar to GB|AAP13431.1|30023796|BT006323 At5g02530 {Ara... 43 1e-04
BP067482 42 3e-04
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 42 4e-04
>TC9308 weakly similar to UP|Q9SQ55 (Q9SQ55) Nuclear RNA binding protein A,
partial (9%)
Length = 562
Score = 180 bits (456), Expect(2) = 6e-67
Identities = 90/94 (95%), Positives = 91/94 (96%)
Frame = +3
Query: 495 YPGEAVEDYMRWYNAVSHRFIIPDDRREEFSAVTVMRRAVDLLEQSLEVSDAPAEGTHSR 554
YPGEAVEDYMRWY+AVSHRFIIPDDRREEFSAVTVMRR VDLLEQSLEV DAPAEGTHSR
Sbjct: 3 YPGEAVEDYMRWYSAVSHRFIIPDDRREEFSAVTVMRRVVDLLEQSLEVPDAPAEGTHSR 182
Query: 555 SLTERALDLIRSNAFIGTQGVAFAAVRGARAAGG 588
SLTERALD IRSNAFIGTQGVAFAAVRGARAAGG
Sbjct: 183 SLTERALDFIRSNAFIGTQGVAFAAVRGARAAGG 284
Score = 92.0 bits (227), Expect(2) = 6e-67
Identities = 46/57 (80%), Positives = 47/57 (81%), Gaps = 10/57 (17%)
Frame = +2
Query: 589 RGRGDRARGGRGRGGRARGEGAPAEGA----------RGGRARGPRGRRGAGRGRGE 635
RGRGDRARGG+GRGGRARGEGAPAEGA RGGRARGPRGRRGAGRGRGE
Sbjct: 284 RGRGDRARGGKGRGGRARGEGAPAEGAPAEGARGGRGRGGRARGPRGRRGAGRGRGE 454
Score = 40.0 bits (92), Expect = 0.001
Identities = 21/32 (65%), Positives = 21/32 (65%)
Frame = +2
Query: 578 AAVRGARAAGGRGRGDRARGGRGRGGRARGEG 609
A GAR GGRGRG RARG RGR G RG G
Sbjct: 362 APAEGAR--GGRGRGGRARGPRGRRGAGRGRG 451
Score = 34.7 bits (78), Expect = 0.048
Identities = 18/36 (50%), Positives = 19/36 (52%)
Frame = +2
Query: 573 QGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGE 608
+G RG R GGR RG R R G GRG RGE
Sbjct: 356 EGAPAEGARGGRGRGGRARGPRGRRGAGRG---RGE 454
>BP066856
Length = 485
Score = 233 bits (595), Expect = 5e-62
Identities = 121/160 (75%), Positives = 131/160 (81%)
Frame = -2
Query: 476 DAAFAEFAPHLRPQGIPATYPGEAVEDYMRWYNAVSHRFIIPDDRREEFSAVTVMRRAVD 535
DAA+A+F PHLRP+GIPATY GE VEDYMRWY+ VSHRFIIPDDRREE SAVTV+RRAVD
Sbjct: 484 DAAYADFVPHLRPRGIPATYSGEXVEDYMRWYSGVSHRFIIPDDRREELSAVTVVRRAVD 305
Query: 536 LLEQSLEVSDAPAEGTHSRSLTERALDLIRSNAFIGTQGVAFAAVRGARAAGGRGRGDRA 595
LLEQSLEV DA A GTH+RSL +RALDLIRS++FIGTQGVAFAAVRGA A GGRGRG RA
Sbjct: 304 LLEQSLEVLDALAVGTHARSLIKRALDLIRSSSFIGTQGVAFAAVRGAGAPGGRGRGGRA 125
Query: 596 RGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRGE 635
RG +GARGGRA GPRGRRGAGRGR E
Sbjct: 124 RG---------------DGARGGRAHGPRGRRGAGRGRVE 50
>BP066148
Length = 477
Score = 184 bits (467), Expect = 4e-47
Identities = 96/102 (94%), Positives = 96/102 (94%)
Frame = -2
Query: 534 VDLLEQSLEVSDAPAEGTHSRSLTERALDLIRSNAFIGTQGVAFAAVRGARAAGGRGRGD 593
VDLLEQSLEVSDAPAEGTHSRSLTERALDLIRSNAFIGTQGVAFAAVRGARAAG RGRGD
Sbjct: 476 VDLLEQSLEVSDAPAEGTHSRSLTERALDLIRSNAFIGTQGVAFAAVRGARAAGXRGRGD 297
Query: 594 RARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRGE 635
RARGGRG RGEGAPAEGARGGRARGPRGRRGAGRGRGE
Sbjct: 296 RARGGRG-----RGEGAPAEGARGGRARGPRGRRGAGRGRGE 186
>BP058107
Length = 496
Score = 179 bits (453), Expect = 2e-45
Identities = 93/128 (72%), Positives = 102/128 (79%), Gaps = 5/128 (3%)
Frame = -3
Query: 510 VSHRFIIPDDRREEFSAVTVMRRAVDLLEQSLEVSDAPAEGTHSRSLTERALDLIRSNAF 569
VSH FIIP+DRREE SAV+ +RR V+LLEQSLEV A A GT R LTERALDL R ++F
Sbjct: 491 VSHVFIIPEDRREELSAVSAIRRGVELLEQSLEVPGAYAPGTQPRILTERALDLFRRSSF 312
Query: 570 IGTQGVAFAAVRGARAAGGRGRGDRARGGR-----GRGGRARGEGAPAEGARGGRARGPR 624
+GTQGVAF+A+RGA AAGGR RG RARGGR RGGRARGEG P EG RGGRARGPR
Sbjct: 311 VGTQGVAFSAIRGAAAAGGRARGGRARGGRPRGGGARGGRARGEGDPGEGVRGGRARGPR 132
Query: 625 GRRGAGRG 632
GRRG GRG
Sbjct: 131 GRRGQGRG 108
>TC19261 similar to UP|BAD06577 (BAD06577) Cell wall protein Awa1p, partial
(3%)
Length = 495
Score = 92.8 bits (229), Expect = 1e-19
Identities = 44/58 (75%), Positives = 47/58 (80%)
Frame = -2
Query: 575 VAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
+AF+A+RGA AAGGR RG R RGG RGGRARGEG P EG RGGRAR PRGRRG GRG
Sbjct: 263 LAFSAIRGAAAAGGRARGGRPRGGGARGGRARGEGDPGEGVRGGRARVPRGRRGRGRG 90
Score = 85.1 bits (209), Expect = 3e-17
Identities = 44/69 (63%), Positives = 51/69 (73%)
Frame = -1
Query: 227 GERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDAARIWLV 286
GERDE AALCAQL+GGSV Y AEF++ QT+RF L+T +A+ RYEDAARIWLV
Sbjct: 495 GERDEVAALCAQLLGGSVAAYLAEFESAGGQTLRFLTLKTMSTSAMDGGRYEDAARIWLV 316
Query: 287 NQLGATLFA 295
N L ATL A
Sbjct: 315 NPLAATLVA 289
Score = 29.6 bits (65), Expect = 1.5
Identities = 17/37 (45%), Positives = 19/37 (50%)
Frame = +1
Query: 589 RGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRG 625
R R R RG R R R G+P+ AR RA PRG
Sbjct: 94 RPRPRRPRGTRARPPRTPSPGSPSPRARPPRAPPPRG 204
>AV769506
Length = 345
Score = 90.5 bits (223), Expect = 7e-19
Identities = 42/42 (100%), Positives = 42/42 (100%)
Frame = -3
Query: 126 YVDRRQLKVATAGGKVWNLACDGDSDSHRRVRELIEQTGLHQ 167
YVDRRQLKVATAGGKVWNLACDGDSDSHRRVRELIEQTGLHQ
Sbjct: 343 YVDRRQLKVATAGGKVWNLACDGDSDSHRRVRELIEQTGLHQ 218
>BP071119
Length = 412
Score = 84.7 bits (208), Expect = 4e-17
Identities = 49/118 (41%), Positives = 63/118 (52%)
Frame = +2
Query: 94 FPGGPVELSLLTHYADHKAPWAWHALLRTDERYVDRRQLKVATAGGKVWNLACDGDSDSH 153
+ GGP +L LLT Y +H A W + R + Y R LK+ G C
Sbjct: 80 YEGGPFDLCLLTKYGEHIARDIWRSYKRPN--YETRGVLKIYNHGRM-----CHPVVHHA 238
Query: 154 RRVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMSFGEMTITLDDVSA 211
R +R + GL + C+ P D +I A VERWH ETSSFH+ +GEMTITL+DVSA
Sbjct: 239 RYIRGAVHNAGLRWVMKCTSPSVDQSIISAFVERWHPETSSFHLPWGEMTITLEDVSA 412
>BP042402
Length = 409
Score = 79.0 bits (193), Expect = 2e-15
Identities = 55/100 (55%), Positives = 63/100 (63%), Gaps = 6/100 (6%)
Frame = -2
Query: 542 EVSDAPAEGTHSRSLTERALDLIRSNAFIGTQGVAFAAVRGARAAGGR--GRGDRARGGR 599
EV DA GT +R+LTERAL +++ A GTQG+A+AA RG AGGR GR GGR
Sbjct: 408 EVDDALLPGTQARALTERALRILKDLA--GTQGIAYAAGRGGLGAGGRAGGRAGGRAGGR 235
Query: 600 GRGGRARG-EGAPAEGAR-GGRARGPRGRRGAG--RGRGE 635
GGRA G EG A G R GGRA GRRG G RGRG+
Sbjct: 234 A-GGRAGGREGGRAAGGREGGRAGARGGRRGRGGRRGRGQ 118
>BP053409
Length = 441
Score = 67.8 bits (164), Expect = 5e-12
Identities = 39/91 (42%), Positives = 51/91 (55%), Gaps = 5/91 (5%)
Frame = -1
Query: 548 AEGTHSRSLTERALDLIRSNAFIGTQGVAFAAVRGARAAGGRGRGDRARGG-----RGRG 602
++G R LTERALDL R ++F+GTQGV F+A+RGA AAG R RG RARG R
Sbjct: 357 SKGHKPRILTERALDLFRRSSFVGTQGVPFSAIRGAAAAGARARGARARGXKTPWRRSSW 178
Query: 603 GRARGEGAPAEGARGGRARGPRGRRGAGRGR 633
++ G G + + +G G GR
Sbjct: 177 SQSSWRGRSWRGXSWXSSSWTQXSQGPGSGR 85
Score = 67.8 bits (164), Expect = 5e-12
Identities = 50/122 (40%), Positives = 59/122 (47%), Gaps = 10/122 (8%)
Frame = -2
Query: 521 REEFSAVTVMRRAVDLLEQSLEVSDAPAEGTHSRSLTERALDLIRSNAFIGTQGVA---- 576
REE SAV+ +RR V+LL++SLEV A A S + R RS +F G +
Sbjct: 440 REELSAVSPLRRGVELLDKSLEVQGAYAPRDTSPESSRRG----RSISFDGVPSLVPREF 273
Query: 577 ------FAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAG 630
+R R RGG RG RARGE P E RG RARGPR RRG
Sbjct: 272 HFLLFEEPQLREPELVEPELVEXRPRGGGARGARARGEVDPGEVVRGXRARGPRXRRGRX 93
Query: 631 RG 632
RG
Sbjct: 92 RG 87
>BP085151
Length = 360
Score = 64.3 bits (155), Expect = 6e-11
Identities = 41/80 (51%), Positives = 46/80 (57%), Gaps = 2/80 (2%)
Frame = -1
Query: 557 TERALDLIRSNAFIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGAR 616
TERAL +++ A GTQG+A+AA RG AGGR G R G GGRA G EG R
Sbjct: 360 TERALRILKDLA--GTQGIAYAAGRGGLGAGGRAGG---RAGVRAGGRAGGRAGGREGGR 196
Query: 617 --GGRARGPRGRRGAGRGRG 634
GGR G G RG RGRG
Sbjct: 195 AAGGREGGRAGARGVRRGRG 136
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 50.4 bits (119), Expect = 8e-07
Identities = 30/55 (54%), Positives = 31/55 (55%), Gaps = 1/55 (1%)
Frame = +1
Query: 581 RGARAAGG-RGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
RG GG RGRGDR RG GGR G G P + ARGG G GR G G GRG
Sbjct: 46 RGRGGGGGFRGRGDRGRGRGDGGGRGGGRGTPFK-ARGGGRGGGGGRGGRGGGRG 207
Score = 27.7 bits (60), Expect = 5.9
Identities = 23/57 (40%), Positives = 27/57 (47%), Gaps = 8/57 (14%)
Frame = +1
Query: 571 GTQGVAFAAVRGARAAGG----RGRGDRARGGRGRGG-RARGEGAPAEG---ARGGR 619
G +G F A G R GG RG G RGG +GG R E EG A+GG+
Sbjct: 124 GGRGTPFKARGGGRGGGGGRGGRGGGRGGRGGGMKGGSRVVVEPHRHEGIFIAKGGK 294
>TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like, complete
Length = 981
Score = 45.8 bits (107), Expect = 2e-05
Identities = 28/54 (51%), Positives = 31/54 (56%), Gaps = 7/54 (12%)
Frame = +2
Query: 588 GRGRGDRARGGRGRGGRARGEGAPAEGARG--GRARG-----PRGRRGAGRGRG 634
GRGRG R G GR + G G +GA+G GR RG P G RGAGRGRG
Sbjct: 386 GRGRGGREEGAGGRPAKGPGRGFD-DGAKGPGGRGRGGLGGKPGGSRGAGRGRG 544
>AV780545
Length = 555
Score = 45.1 bits (105), Expect = 4e-05
Identities = 23/49 (46%), Positives = 30/49 (60%)
Frame = +1
Query: 158 ELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMSFGEMTITL 206
E +E+ G L D LI ALVERW ET++FH++ GEMT+TL
Sbjct: 409 E*VERAGFGYLRSIPAISLDNPLISALVERWRRETNTFHLNVGEMTVTL 555
>TC8572 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 homolog
(At3g57490), partial (91%)
Length = 1076
Score = 44.7 bits (104), Expect = 5e-05
Identities = 29/64 (45%), Positives = 31/64 (48%)
Frame = +2
Query: 568 AFIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRR 627
+F+ F A R G RG GDR GRG GGR RG G R RG R RR
Sbjct: 26 SFLSAPHSLFTASMAERGGGERGGGDRGGFGRGFGGRGRG---------GDRGRGGR-RR 175
Query: 628 GAGR 631
GAGR
Sbjct: 176 GAGR 187
Score = 32.7 bits (73), Expect = 0.18
Identities = 17/26 (65%), Positives = 17/26 (65%), Gaps = 1/26 (3%)
Frame = +2
Query: 584 RAAGGRGRG-DRARGGRGRGGRARGE 608
R GGRGRG DR RGGR RG R E
Sbjct: 119 RGFGGRGRGGDRGRGGRRRGAGRRDE 196
>TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%)
Length = 562
Score = 43.9 bits (102), Expect = 8e-05
Identities = 31/77 (40%), Positives = 36/77 (46%), Gaps = 3/77 (3%)
Frame = +2
Query: 559 RALDLIRSNAFIGTQGVAFAAVRGARAAGGR---GRGDRARGGRGRGGRARGEGAPAEGA 615
+AL+L S+ T V A R + +GGR GR RGGR GR G G G
Sbjct: 59 KALELHDSDLGGFTLAVDEAKPRDNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGG 238
Query: 616 RGGRARGPRGRRGAGRG 632
RGG G G G GRG
Sbjct: 239 RGGDRGGRGGSGGGGRG 289
Score = 38.9 bits (89), Expect = 0.003
Identities = 27/54 (50%), Positives = 28/54 (51%), Gaps = 3/54 (5%)
Frame = +2
Query: 582 GARAAGGR---GRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
G R+ GGR GRG R GRG GGR R G RGGR G RG GRG
Sbjct: 143 GGRSGGGRSGGGRGGRFDSGRG-GGRGRFGSGGRGGDRGGRGGSGGGGRG-GRG 298
Score = 37.0 bits (84), Expect = 0.010
Identities = 19/39 (48%), Positives = 19/39 (48%)
Frame = +2
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGR 619
RG R GRG G G GRGG G G G RGGR
Sbjct: 179 RGGRFDSGRGGGRGRFGSGGRGGDRGGRGGSGGGGRGGR 295
Score = 36.2 bits (82), Expect = 0.016
Identities = 28/57 (49%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Frame = +2
Query: 582 GARAAGGRG-RGDRARGG-RGR---GGRARGEGAPAEGARGGRARGPRGR--RGAGR 631
G R+ GGRG R D RGG RGR GGR G G GG RG RG R AG+
Sbjct: 158 GGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRG--GRGGSGGGGRGGRGTPLRAAGK 322
>TC8182 weakly similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, partial
(13%)
Length = 611
Score = 43.1 bits (100), Expect = 1e-04
Identities = 34/85 (40%), Positives = 37/85 (43%), Gaps = 15/85 (17%)
Frame = +3
Query: 565 RSNAFIGTQGVAFAAVRGARAAGGRGRGDRARG-GRGRGGRARGEGAPAEGAR------- 616
R F G+ + G GGRG G R RG GRGRG RG G GA+
Sbjct: 3 RGRGFYNNGGMEYGG--GDGWDGGRGYGGRGRGFGRGRGRGFRGRGRGGYGAQPVGYYDY 176
Query: 617 -------GGRARGPRGRRGAGRGRG 634
R RG RGR G GRGRG
Sbjct: 177 GEYDAPPAPRGRG-RGRGGRGRGRG 248
Score = 29.6 bits (65), Expect = 1.5
Identities = 21/58 (36%), Positives = 21/58 (36%), Gaps = 26/58 (44%)
Frame = +3
Query: 581 RGARAAGGRGRGDRARG--------------------------GRGRGGRARGEGAPA 612
RG GRGRG R RG GRGRGGR RG G A
Sbjct: 84 RGRGFGRGRGRGFRGRGRGGYGAQPVGYYDYGEYDAPPAPRGRGRGRGGRGRGRGRNA 257
>BI420016
Length = 559
Score = 43.1 bits (100), Expect = 1e-04
Identities = 28/59 (47%), Positives = 29/59 (48%), Gaps = 6/59 (10%)
Frame = -1
Query: 581 RGARAAGGRGRGDRARGGRGRGGRA------RGEGAPAEGARGGRARGPRGRRGAGRGR 633
RG R GG R R R GRGGR R + G RGGR RG R RRG RGR
Sbjct: 445 RGQRGGGGGQRRGRCRRASGRGGRIGSRRGRRRRRSRRRGRRGGRERGVRSRRGR-RGR 272
Score = 42.7 bits (99), Expect = 2e-04
Identities = 30/61 (49%), Positives = 31/61 (50%), Gaps = 8/61 (13%)
Frame = -1
Query: 581 RGARAAGGRGRGDRARGGR-----GRGGRARGEGAPAEGARGGRA---RGPRGRRGAGRG 632
R GGRGRG R GGR G GG+ RG A G RGGR RG R RR RG
Sbjct: 502 RRGGGGGGRGRGRREEGGRRGQRGGGGGQRRGRCRRASG-RGGRIGSRRGRRRRRSRRRG 326
Query: 633 R 633
R
Sbjct: 325 R 323
Score = 36.6 bits (83), Expect = 0.013
Identities = 22/46 (47%), Positives = 24/46 (51%)
Frame = -1
Query: 587 GGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
GG R GGRGRG R G G GG+ RG R RR +GRG
Sbjct: 514 GGCCRRGGGGGGRGRGRREEGGRRGQRGGGGGQRRG-RCRRASGRG 380
Score = 32.7 bits (73), Expect = 0.18
Identities = 26/59 (44%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Frame = -1
Query: 578 AAVRGARAAGGRGRGDRARGGRG-RGGRARGEGAPAEGARGGRARGPRG-RRGAGRGRG 634
A+ RG R RGR R RG RGGR RG + G RG R+ G RG+G G G
Sbjct: 394 ASGRGGRIGSRRGRRRRRSRRRGRRGGRERGVRS-RRGRRGRRSESWGGWWRGSGCGNG 221
>TC8052 similar to GB|AAP13431.1|30023796|BT006323 At5g02530 {Arabidopsis
thaliana;}, partial (55%)
Length = 1389
Score = 43.1 bits (100), Expect = 1e-04
Identities = 50/164 (30%), Positives = 65/164 (39%), Gaps = 21/164 (12%)
Frame = +2
Query: 493 ATYPGE------AVEDYMRWYNAVSHRFIIPDDRREEFSAVTVMRRAVDLLEQSLEVSDA 546
A +PG ++E + Y + + DD +E FS V ++R ++S S
Sbjct: 350 AAFPGHGGGRASSIETGTKLYISNLDYGVSSDDIKELFSEVGDLKRHTVHYDRSGR-SKG 526
Query: 547 PAEGTHSRSLTERALDLIRSNA----------FIGTQGVAFAAVRGARAAGGRGRGDRAR 596
AE SR A +N +GT A A A G G R
Sbjct: 527 TAEVVFSRRGDAEAAVKRYNNVQLDGKPMKIEIVGTNITTPAVAPAANGAFGNSNGV-PR 703
Query: 597 GGRGRGG-----RARGEGAPAEGARGGRARGPRGRRGAGRGRGE 635
G+GRGG R G+G G R GR RG RG GRGRGE
Sbjct: 704 SGQGRGGAFGRLRGGGDGGRGRGHRRGRGRG----RGHGRGRGE 823
Score = 30.4 bits (67), Expect = 0.90
Identities = 21/47 (44%), Positives = 25/47 (52%), Gaps = 6/47 (12%)
Frame = +2
Query: 573 QGVAFAAVRGARAAGGRGRGDR-----ARG-GRGRGGRARGEGAPAE 613
+G AF +RG GGRGRG R RG GRGRG + E A+
Sbjct: 716 RGGAFGRLRGG-GDGGRGRGHRRGRGRGRGHGRGRGEKVSAEDLDAD 853
>BP067482
Length = 291
Score = 42.0 bits (97), Expect = 3e-04
Identities = 28/87 (32%), Positives = 41/87 (46%), Gaps = 1/87 (1%)
Frame = +2
Query: 221 FYTPGRGERDECAALCAQLMGGSVG-IYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYED 279
F++ G R+E A AQL+ + I EFD R ++RF LQ E + +
Sbjct: 23 FFSFGNPTREEAAPAVAQLLDVDIETI**EEFDACRGPSLRFSFLQAIVETHVEANNEVP 202
Query: 280 AARIWLVNQLGATLFASKSGGYHTTVY 306
A R +++ +G TL KS Y VY
Sbjct: 203 ACRAYMLRWIGMTLVCDKSNTYIGAVY 283
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 41.6 bits (96), Expect = 4e-04
Identities = 42/142 (29%), Positives = 57/142 (39%), Gaps = 7/142 (4%)
Frame = +3
Query: 500 VEDYMRWYN-AVSHRFIIPDDRREEFSA--VTVMRRAVDLLEQSLEVSDAPAEGTHSRSL 556
V ++W+N FI PDD ++ + L + +V A R+
Sbjct: 48 VNGKVKWFNDQKGFGFITPDDGGDDLFVHQSEIKSEGFRCLGEGEDVEFIVASDPDGRA- 224
Query: 557 TERALDLIRSN--AFIGTQ--GVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPA 612
+A+++ N A GT+ G A RG GG G G GG GRGG G G
Sbjct: 225 --KAIEVTGPNGQAVQGTRRGGGAGGGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGGGGY 398
Query: 613 EGARGGRARGPRGRRGAGRGRG 634
G+ G G G G G GRG
Sbjct: 399 GGSGGYGGGGGYGGGGRGGGRG 464
Score = 41.2 bits (95), Expect = 5e-04
Identities = 23/53 (43%), Positives = 23/53 (43%)
Frame = +3
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G GG G G GGRG GG G G G GG G RG GRG G
Sbjct: 312 GGYGGGGYGGGGYGGGGRGGGGYGGGGGYGGSGGYGGGGGYGGGGRGGGRGGG 470
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,485,664
Number of Sequences: 28460
Number of extensions: 153858
Number of successful extensions: 2042
Number of sequences better than 10.0: 188
Number of HSP's better than 10.0 without gapping: 1467
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1781
length of query: 635
length of database: 4,897,600
effective HSP length: 96
effective length of query: 539
effective length of database: 2,165,440
effective search space: 1167172160
effective search space used: 1167172160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0042.1


