

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
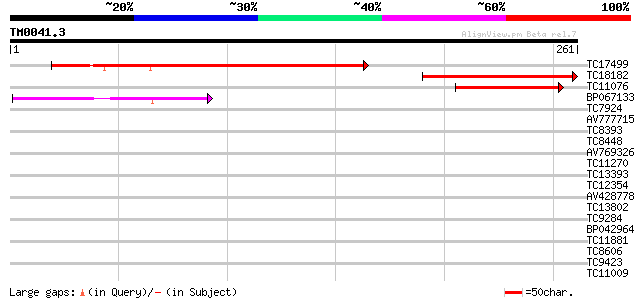
Query= TM0041.3
(261 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17499 weakly similar to UP|O82212 (O82212) At2g23820 protein, ... 152 4e-38
TC18182 similar to UP|O82212 (O82212) At2g23820 protein, partial... 139 4e-34
TC11076 77 2e-15
BP067133 40 4e-04
TC7924 homologue to UP|CAE53883 (CAE53883) Aquaporin, partial (95%) 38 0.002
AV777715 38 0.002
TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenyla... 37 0.003
TC8448 weakly similar to UP|WSC4_YEAST (P38739) Cell wall integr... 37 0.003
AV769326 37 0.004
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 36 0.006
TC13393 similar to UP|Q9FMU6 (Q9FMU6) Mitochondrial phosphate tr... 36 0.006
TC12354 similar to UP|O23702 (O23702) Dehydrogenase, partial (25%) 35 0.010
AV428778 35 0.010
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 35 0.010
TC9284 weakly similar to UP|Q9LPY0 (Q9LPY0) T23J18.22, partial (... 35 0.010
BP042964 35 0.010
TC11881 similar to PIR|T48303|T48303 meiosis specific-like prote... 35 0.010
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 35 0.013
TC9423 similar to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein... 35 0.013
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 35 0.017
>TC17499 weakly similar to UP|O82212 (O82212) At2g23820 protein, partial
(40%)
Length = 502
Score = 152 bits (385), Expect = 4e-38
Identities = 80/157 (50%), Positives = 111/157 (69%), Gaps = 11/157 (7%)
Frame = +2
Query: 20 SLSSTQLRFKSVTLSPHSPSRVF----HMTTAAGSSSPPSSSVTTSPP-------SAASA 68
S+SS+ K + H +R++ H ++++ S+ P SP SA
Sbjct: 35 SVSSSSFSLKMAAIHSHI-ARLWVPPLHRSSSSPSNHPKLLLFRHSPSLSLRAHSSAEQH 211
Query: 69 TPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDK 128
+ S+SS IDFL++C+RLKTTKR GW+ ++ ESIADHMYRM++MALIAAD+PG+ R++
Sbjct: 212 SHSSSSVIDFLTLCNRLKTTKRKGWVNHGIKGAESIADHMYRMAIMALIAADVPGLSRER 391
Query: 129 CVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEAL 165
C+K+A+VHDIAEAIVGDITP+DGVPK EK+R+EQEAL
Sbjct: 392 CIKIALVHDIAEAIVGDITPSDGVPKAEKSRMEQEAL 502
>TC18182 similar to UP|O82212 (O82212) At2g23820 protein, partial (19%)
Length = 556
Score = 139 bits (351), Expect = 4e-34
Identities = 68/71 (95%), Positives = 69/71 (96%)
Frame = +2
Query: 191 NSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
NSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFF STAGKF TEIG+AWASEIVSRRK
Sbjct: 2 NSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFPSTAGKFPTEIGQAWASEIVSRRK 181
Query: 251 KTNGSSHPHST 261
KTNGSSHPHST
Sbjct: 182 KTNGSSHPHST 214
>TC11076
Length = 481
Score = 77.4 bits (189), Expect = 2e-15
Identities = 38/50 (76%), Positives = 42/50 (84%)
Frame = +3
Query: 206 EMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKKTNGS 255
EMIL ALEYE E GK LDEFF STAGKFQTEIGK+WASEI+SRRK + +
Sbjct: 3 EMILPALEYEMEPGKVLDEFFLSTAGKFQTEIGKSWASEIISRRKSLSAN 152
>BP067133
Length = 507
Score = 40.0 bits (92), Expect = 4e-04
Identities = 33/95 (34%), Positives = 45/95 (46%), Gaps = 3/95 (3%)
Frame = +1
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
A+ T +S ++S +S ++T S T SP SPS +SSPPS+ TS
Sbjct: 244 ASTTTASSASSSATSTSTSPTTTTTATSSPTASPSSPS-------PTPASSPPSAPSRTS 402
Query: 62 PPS---AASATPSASSAIDFLSICHRLKTTKRAGW 93
PPS + + TPSASS I S + R W
Sbjct: 403 PPSIPFSNTWTPSASSLISGPSTSTSHYSAARIAW 507
>TC7924 homologue to UP|CAE53883 (CAE53883) Aquaporin, partial (95%)
Length = 1078
Score = 38.1 bits (87), Expect = 0.002
Identities = 29/74 (39%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Frame = +3
Query: 11 SLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPS--AASA 68
SL SSS PSS S++ S T S + V + G S+ SSS +T+PP+ AA++
Sbjct: 132 SLPSSSPPSSSSTSPSPPSSATRSKPDHATVSASSALPGPSAA*SSSSSTAPPASPAATS 311
Query: 69 TPSASSAIDFLSIC 82
TP + SA L+ C
Sbjct: 312 TPPSPSASSSLARC 353
>AV777715
Length = 398
Score = 37.7 bits (86), Expect = 0.002
Identities = 24/66 (36%), Positives = 34/66 (51%)
Frame = +2
Query: 13 FSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSA 72
F S SSL + R+ TLSP SP+R H ++ S + SS + PS S +P++
Sbjct: 92 FPSPCASSLVPSSTRYS--TLSPDSPTRRVHSMASSSSRARSSSPFSHRKPSTPSYSPAS 265
Query: 73 SSAIDF 78
SS F
Sbjct: 266 SSYSSF 283
>TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenylated Rab
receptor 2 {Arabidopsis thaliana;}, partial (78%)
Length = 1012
Score = 37.0 bits (84), Expect = 0.003
Identities = 23/66 (34%), Positives = 37/66 (55%)
Frame = +1
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS S + ++ S + + +S+ ++PHSP+R S PPS+S SP SA++ T
Sbjct: 166 SSTTSPNPSATASLSAVPGRSLPIAPHSPNR-------NPSPMPPSASARISPTSASTTT 324
Query: 70 PSASSA 75
PS S+
Sbjct: 325 PSFRSS 342
Score = 26.9 bits (58), Expect = 3.5
Identities = 25/80 (31%), Positives = 35/80 (43%), Gaps = 14/80 (17%)
Frame = +3
Query: 46 TAAGSSSPPSSSVTTSPP------SAASATPSASSA----IDFLSICHRLKTTKRAGWI- 94
T S++PP V+ +PP SA S P+ S A I+ LS R ++R W
Sbjct: 48 TTMSSTAPPVLPVSNTPPASTTVASAGSDAPATSPAFRALINNLSESLRNGLSQRRPWSE 227
Query: 95 ---RKDVQDPESIADHMYRM 111
R PES +D R+
Sbjct: 228 LTDRSAFSKPESFSDATLRV 287
>TC8448 weakly similar to UP|WSC4_YEAST (P38739) Cell wall integrity and
stress response component 4 precursor, partial (5%)
Length = 1049
Score = 37.0 bits (84), Expect = 0.003
Identities = 23/62 (37%), Positives = 34/62 (54%)
Frame = +1
Query: 13 FSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSA 72
F S+ P SLS R H P RVF+ + + SSSP S++ ++SP S SA+ S
Sbjct: 61 FISASPPSLSLQSHR------PSHLPRRVFYPSASLSSSSPESTASSSSPQSGPSASTST 222
Query: 73 SS 74
++
Sbjct: 223 TT 228
>AV769326
Length = 262
Score = 36.6 bits (83), Expect = 0.004
Identities = 26/60 (43%), Positives = 30/60 (49%)
Frame = +2
Query: 16 SIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
S PS S S P+SPS TA PPSS +T PPS +S+TPSA SA
Sbjct: 83 SPPSDPHSPPSNPPSAASDPNSPS-----LTAPSLQKPPSSPTSTEPPS-SSSTPSAPSA 244
Score = 26.9 bits (58), Expect = 3.5
Identities = 19/52 (36%), Positives = 25/52 (47%)
Frame = +2
Query: 16 SIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAAS 67
S S +S L S+ P SP+ S+ PPSSS T S PSA++
Sbjct: 125 SAASDPNSPSLTAPSLQKPPSSPT----------STEPPSSSSTPSAPSAST 250
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 36.2 bits (82), Expect = 0.006
Identities = 25/57 (43%), Positives = 34/57 (58%)
Frame = -3
Query: 19 SSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
S L ST L F S T +P S S +++ SS P SSS TTS S++S+ S+SS+
Sbjct: 580 SPLESTFLPFTSTTTTPSSSS------SSSSSSFPCSSSTTTSSSSSSSSPFSSSSS 428
>TC13393 similar to UP|Q9FMU6 (Q9FMU6) Mitochondrial phosphate translocator
(AT5g14040/MUA22_4), partial (8%)
Length = 541
Score = 36.2 bits (82), Expect = 0.006
Identities = 26/69 (37%), Positives = 36/69 (51%), Gaps = 3/69 (4%)
Frame = +1
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAA--- 66
SS F + P+ SS SV P +PS+ F +T ++P +SS TSPPS A
Sbjct: 70 SSTFPNPPPTRTSSPSSSPHSVP-PPSTPSKSFPITPTGSLAAPAASSSRTSPPSPAPSL 246
Query: 67 SATPSASSA 75
S P +SS+
Sbjct: 247 SPNPRSSSS 273
Score = 27.3 bits (59), Expect = 2.7
Identities = 24/66 (36%), Positives = 30/66 (45%), Gaps = 4/66 (6%)
Frame = +1
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAAS----AT 69
+S SS S Q F+ V +P S S F +SSP SS + PPS S T
Sbjct: 1 ASLASSSSSPCQWGFRLV--NPPSASSTFPNPPPTRTSSPSSSPHSVPPPSTPSKSFPIT 174
Query: 70 PSASSA 75
P+ S A
Sbjct: 175PTGSLA 192
Score = 26.6 bits (57), Expect = 4.6
Identities = 18/57 (31%), Positives = 27/57 (46%)
Frame = +1
Query: 19 SSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
+S S T + +LSP+ S T++A +P SS P+ S TPS+ A
Sbjct: 202 ASSSRTSPPSPAPSLSPNPRSSSSSPTSSASPKTPMISSPGLRSPNTDSTTPSSMQA 372
>TC12354 similar to UP|O23702 (O23702) Dehydrogenase, partial (25%)
Length = 488
Score = 35.4 bits (80), Expect = 0.010
Identities = 34/99 (34%), Positives = 46/99 (46%), Gaps = 8/99 (8%)
Frame = +3
Query: 3 TATRVLLSSLFSS--SIPSSLS----STQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSS 56
TA+++ SS S S PSS S S R LS +PS FH +A S+ SS
Sbjct: 36 TASKIAPSSSNPSPASPPSSTSLSAVSPMARSSPPPLSSSTPSPTFHAPPSAVSALTTSS 215
Query: 57 SVTTSPPSAASATPS--ASSAIDFLSICHRLKTTKRAGW 93
S ++PP+A S PS S++ F S H + W
Sbjct: 216 S-ASAPPTAPSTLPSLLTSASASFTSTPHAPMRSLTPSW 329
>AV428778
Length = 323
Score = 35.4 bits (80), Expect = 0.010
Identities = 16/39 (41%), Positives = 24/39 (61%)
Frame = +2
Query: 35 PHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
PHSP+R H T ++ +S+ SS + P A+ TPS+S
Sbjct: 14 PHSPTRTPHTTASSSTSAESSSRPHSQPSGPAAQTPSSS 130
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 35.4 bits (80), Expect = 0.010
Identities = 26/73 (35%), Positives = 37/73 (50%)
Frame = +2
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
++T + S+ S S P++ SS+ + S + SR + A S SPPSS SP
Sbjct: 284 SSTPLTPSTATSESSPTATSSSSSASPAPPTSFSALSRALELKALASSPSPPSSP-PPSP 460
Query: 63 PSAASATPSASSA 75
PSA + S SSA
Sbjct: 461 PSAICMSTSLSSA 499
Score = 33.1 bits (74), Expect = 0.050
Identities = 26/64 (40%), Positives = 33/64 (50%), Gaps = 7/64 (10%)
Frame = +2
Query: 18 PSSLSSTQLRFKSVTLSPHS------PSRVFHMTTAAGSSSPPSSSVTTSP-PSAASATP 70
PS +SS + S T SP S PS T A SSSP S+S +SP S ++P
Sbjct: 86 PSQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPASSPTKSPKPSSP 265
Query: 71 SASS 74
SAS+
Sbjct: 266 SASA 277
>TC9284 weakly similar to UP|Q9LPY0 (Q9LPY0) T23J18.22, partial (33%)
Length = 898
Score = 35.4 bits (80), Expect = 0.010
Identities = 34/93 (36%), Positives = 47/93 (49%), Gaps = 15/93 (16%)
Frame = +1
Query: 18 PSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPS---AASAT----- 69
PSS SST L S T +P+S S F T++ SS PPSS T +PP ++SAT
Sbjct: 274 PSSSSSTPL---SSTTNPNSASPSF--ATSSPSSPPPSSPTTPTPPHSRISSSATLNSRN 438
Query: 70 -----PSASSAIDFLSICHRLKTTKR--AGWIR 95
P+++S + S L T R + WI+
Sbjct: 439 RSLVSPNSASMLSPTSASWVLTTVSRTPSRWIK 537
Score = 27.7 bits (60), Expect = 2.1
Identities = 25/88 (28%), Positives = 38/88 (42%), Gaps = 17/88 (19%)
Frame = +1
Query: 1 MATATRVLLSSLFSSSIPS---SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSS 57
++T T S+ FS S P + S++ + + T P P H T + SSS P SS
Sbjct: 139 LSTPTTTNESASFSPSNPDPNPASSASTTKPSAATSPPSKP----HAPTPSSSSSTPLSS 306
Query: 58 VT--------------TSPPSAASATPS 71
T +SPP ++ TP+
Sbjct: 307 TTNPNSASPSFATSSPSSPPPSSPTTPT 390
>BP042964
Length = 575
Score = 35.4 bits (80), Expect = 0.010
Identities = 26/68 (38%), Positives = 35/68 (51%), Gaps = 5/68 (7%)
Frame = +2
Query: 13 FSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSV---TTSPPSAAS-- 67
+SSS P S SS T +PHS + T A S PP++S TT PPS A
Sbjct: 35 WSSSPPPSSSSPTAAS*PATSNPHSNASSAASTAADSSPPPPATSTPSNTTQPPSTARTF 214
Query: 68 ATPSASSA 75
+ P+AS++
Sbjct: 215 SPPTASTS 238
>TC11881 similar to PIR|T48303|T48303 meiosis specific-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(34%)
Length = 631
Score = 35.4 bits (80), Expect = 0.010
Identities = 27/67 (40%), Positives = 36/67 (53%), Gaps = 10/67 (14%)
Frame = +3
Query: 19 SSLSSTQLRFKS--VTLSPHS------PSRVFHMTTAAGSS--SPPSSSVTTSPPSAASA 68
SSLS + R S VT SP P+ + + A SS +PPS+ + TSPPSA
Sbjct: 297 SSLSPARSRTPSSPVTASPSMSPPAPPPTSSMSLNSTASSSRINPPSARLVTSPPSANPP 476
Query: 69 TPSASSA 75
+P ASS+
Sbjct: 477 SPPASSS 497
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 35.0 bits (79), Expect = 0.013
Identities = 26/68 (38%), Positives = 36/68 (52%), Gaps = 4/68 (5%)
Frame = +1
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP----PSA 65
SS SSS SSLSS+ S + P S S ++++ S S PSSS ++SP P+
Sbjct: 277 SSSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSGPAP 456
Query: 66 ASATPSAS 73
A PS +
Sbjct: 457 APGPPSGA 480
Score = 30.4 bits (67), Expect = 0.32
Identities = 24/63 (38%), Positives = 36/63 (57%)
Frame = +1
Query: 11 SLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATP 70
S SSS SS SS S + +P S S ++++ SSS SSS +SP S++S++P
Sbjct: 271 SASSSSSSSSSSSL-----SSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSP 435
Query: 71 SAS 73
+S
Sbjct: 436 FSS 444
Score = 29.6 bits (65), Expect = 0.55
Identities = 15/31 (48%), Positives = 26/31 (83%)
Frame = +1
Query: 45 TTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
+++ SSS PSSS ++SPPS++S++ S+SS+
Sbjct: 301 SSSLSSSSTPSSS-SSSPPSSSSSSSSSSSS 390
Score = 26.2 bits (56), Expect = 6.1
Identities = 14/39 (35%), Positives = 24/39 (60%)
Frame = +1
Query: 37 SPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
S + V+ G S+ SSS ++S ++S+TPS+SS+
Sbjct: 229 SANVVYFFLFLGGCSASSSSSSSSSSSLSSSSTPSSSSS 345
Score = 26.2 bits (56), Expect = 6.1
Identities = 14/33 (42%), Positives = 25/33 (75%), Gaps = 2/33 (6%)
Frame = +1
Query: 45 TTAAGSSSPPSSSVTTS--PPSAASATPSASSA 75
+ ++ SSS SSS+++S P S++S+ PS+SS+
Sbjct: 271 SASSSSSSSSSSSLSSSSTPSSSSSSPPSSSSS 369
>TC9423 similar to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein B13-1-9,
partial (53%)
Length = 969
Score = 35.0 bits (79), Expect = 0.013
Identities = 26/73 (35%), Positives = 40/73 (54%)
Frame = +2
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
++A+ LSS SSS PS SS S +LS H+PS T + SS+ P + TT+
Sbjct: 167 SSASSGRLSSQSSSSSPSYSSS------STSLSNHAPSSSMSPTPNSHSSTTPPPTTTTA 328
Query: 62 PPSAASATPSASS 74
++ + T S++S
Sbjct: 329 TTTSFATTLSSTS 367
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 34.7 bits (78), Expect = 0.017
Identities = 27/73 (36%), Positives = 37/73 (49%)
Frame = -2
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
AT R S SSS SS SS+ S + SP P +++ SSS SSS
Sbjct: 267 ATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPPPG 88
Query: 62 PPSAASATPSASS 74
S++S++PS+SS
Sbjct: 87 ASSSSSSSPSSSS 49
Score = 33.5 bits (75), Expect = 0.038
Identities = 24/67 (35%), Positives = 38/67 (55%), Gaps = 1/67 (1%)
Frame = -2
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPP-SSSVTTSPPSAASA 68
SS SSS SS SS+ P +P ++++ SS PP +SS ++S PS++S
Sbjct: 225 SSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPPPGASSSSSSSPSSSSP 46
Query: 69 TPSASSA 75
+ S+SS+
Sbjct: 45 SSSSSSS 25
Score = 28.9 bits (63), Expect = 0.93
Identities = 17/61 (27%), Positives = 32/61 (51%)
Frame = -2
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
SSS +++ T+ + S S S ++++ S PP +T P S++S++ S+S
Sbjct: 282 SSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSS 103
Query: 74 S 74
S
Sbjct: 102 S 100
Score = 27.3 bits (59), Expect = 2.7
Identities = 25/70 (35%), Positives = 38/70 (53%), Gaps = 3/70 (4%)
Frame = -2
Query: 9 LSSLFSSSIPSSL-SSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS--PPSA 65
LSS SSS SL +S+ +V + +S + ++++ SSS SSS + S PP
Sbjct: 330 LSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRP 151
Query: 66 ASATPSASSA 75
S PS+SS+
Sbjct: 150 PSTPPSSSSS 121
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,376,508
Number of Sequences: 28460
Number of extensions: 62631
Number of successful extensions: 1891
Number of sequences better than 10.0: 387
Number of HSP's better than 10.0 without gapping: 1325
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1666
length of query: 261
length of database: 4,897,600
effective HSP length: 88
effective length of query: 173
effective length of database: 2,393,120
effective search space: 414009760
effective search space used: 414009760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0041.3


