

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0040.4
(457 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
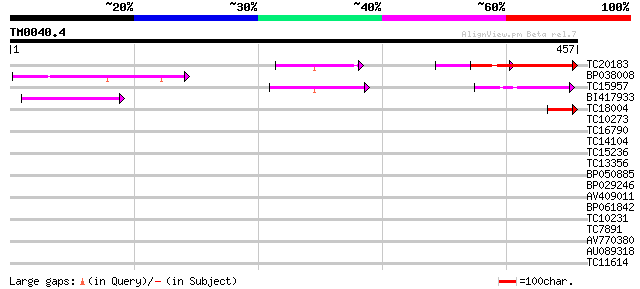
Score E
Sequences producing significant alignments: (bits) Value
TC20183 weakly similar to UP|Q9LHY9 (Q9LHY9) ESTs C74322(E30878)... 136 8e-33
BP038008 57 5e-09
TC15957 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization resp... 52 2e-07
BI417933 45 3e-05
TC18004 44 7e-05
TC10273 35 0.026
TC16790 32 0.17
TC14104 GB|CAA61590.1|897773|LJAS2GENE asparagine synthase (glut... 31 0.49
TC15236 homologue to UP|GCST_PEA (P49364) Aminomethyltransferase... 29 1.4
TC13356 weakly similar to UP|Q9SVG3 (Q9SVG3) Reticuline oxidase-... 29 1.8
BP050885 28 3.2
BP029246 28 3.2
AV409011 28 3.2
BP061842 28 4.1
TC10231 similar to UP|O23110 (O23110) AP2 domain containing prot... 27 5.4
TC7891 similar to UP|DCAM_VICFA (Q9M4D8) S-adenosylmethionine de... 27 5.4
AV770380 27 5.4
AU089318 27 7.0
TC11614 similar to UP|Q9FHH4 (Q9FHH4) Diacylglycerol kinase-like... 27 9.2
>TC20183 weakly similar to UP|Q9LHY9 (Q9LHY9) ESTs C74322(E30878), partial
(14%)
Length = 516
Score = 136 bits (342), Expect = 8e-33
Identities = 66/86 (76%), Positives = 74/86 (85%)
Frame = +3
Query: 372 RPRVPKLFIRKHFNDIQQSEQTVILQCEKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKL 431
RPRVP FIR++FND+QQ T++LQ EKKL PVKL CYPKKG +LS+GWVEFA KSKL
Sbjct: 90 RPRVPNPFIREYFNDMQQ---TIMLQYEKKLWPVKLICYPKKGSGKLSQGWVEFAGKSKL 260
Query: 432 EVGDACVFELINKEDPVLDVHIYRGH 457
VGDACVFELINKEDPVLDVHI+RGH
Sbjct: 261 VVGDACVFELINKEDPVLDVHIFRGH 338
Score = 59.3 bits (142), Expect = 1e-09
Identities = 31/64 (48%), Positives = 38/64 (58%)
Frame = +2
Query: 344 AALEEANKFNSNNPFFIVNIAPDKNRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEKKLC 403
AALEEANKFNSNNPFFIVN+APD DYRP + + ++ V ++ E C
Sbjct: 2 AALEEANKFNSNNPFFIVNLAPDMEWDYRPTACTKSLHQRVLQ*HAADHNVTVRKEIVAC 181
Query: 404 PVKL 407
V L
Sbjct: 182 EVDL 193
Score = 41.6 bits (96), Expect = 3e-04
Identities = 25/78 (32%), Positives = 39/78 (49%), Gaps = 7/78 (8%)
Frame = +3
Query: 215 IPSFFAKKYLKNETDVLLQVLDGRTWSVSF------NLGKFNAGWKKFTSVNNLKVGDVC 268
+P+ F ++Y + ++ + + W V GK + GW +F + L VGD C
Sbjct: 99 VPNPFIREYFNDMQQTIMLQYEKKLWPVKLICYPKKGSGKLSQGWVEFAGKSKLVVGDAC 278
Query: 269 LFEL-NKSVPLSLKVLIF 285
+FEL NK P+ L V IF
Sbjct: 279 VFELINKEDPV-LDVHIF 329
>BP038008
Length = 509
Score = 57.4 bits (137), Expect = 5e-09
Identities = 42/153 (27%), Positives = 73/153 (47%), Gaps = 10/153 (6%)
Frame = +2
Query: 3 MESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDV-WFQNGWKDFAKYYS 61
+++ V R+P ++ ++NP L P G W++ K DV + WK+FA +YS
Sbjct: 11 LQNGVLRMPRKLVSKHWDGISNPISLLLP-GATWEVRWEKRGYDVIMLGDEWKNFAMHYS 187
Query: 62 LDHGHMVLFEYKDTSH-----FGVHIFDKSTLEIRYPSHE-NQDEEHNPDQIDDDSVEIL 115
LD H++ F ++ SH F V I + YP + + D D+ DD E
Sbjct: 188 LDVEHLLRFSFRHDSHVRRTQFHVEIHESGMEIDHYPFKQGSADSGEESDKSTDDENENE 367
Query: 116 DKIPPC---KKTKLKSPMSCPKPRKKLRTSTSE 145
++ +K + KSP+ + KK++T++ E
Sbjct: 368 KEVHVSSRHQKKRPKSPLPVSRSPKKVKTNSKE 466
>TC15957 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization response 1,
partial (26%)
Length = 627
Score = 52.4 bits (124), Expect = 2e-07
Identities = 33/86 (38%), Positives = 48/86 (55%), Gaps = 5/86 (5%)
Frame = +1
Query: 210 GYSLHIPSFFAKKYLKNETDVL-LQVLDGRTWSVSF----NLGKFNAGWKKFTSVNNLKV 264
G +++PS FA+K L + + LQV DGR V K + GW +F N+L
Sbjct: 13 GCIMYLPSCFAEKNLNGXSGFIKLQVADGRQXPVRCLYRGGRAKLSQGWFEFPLENHLGE 192
Query: 265 GDVCLFELNKSVPLSLKVLIFPLAEE 290
GDVC+FEL ++ + L+V +F L EE
Sbjct: 193 GDVCVFELLRTRDVVLQVTLFRLTEE 270
Score = 47.8 bits (112), Expect = 4e-06
Identities = 27/81 (33%), Positives = 43/81 (52%)
Frame = +1
Query: 375 VPKLFIRKHFNDIQQSEQTVILQCEKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVG 434
+P F K+ N + + + + PV+ C + G ++LS+GW EF ++ L G
Sbjct: 28 LPSCFAEKNLNGXSGFIKLQV--ADGRQXPVR--CLYRGGRAKLSQGWFEFPLENHLGEG 195
Query: 435 DACVFELINKEDPVLDVHIYR 455
D CVFEL+ D VL V ++R
Sbjct: 196 DVCVFELLRTRDVVLQVTLFR 258
>BI417933
Length = 568
Score = 44.7 bits (104), Expect = 3e-05
Identities = 24/83 (28%), Positives = 39/83 (46%)
Frame = +2
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F + G ++ N L+ P G + + + GD G + + + H MV
Sbjct: 251 VPKRFHEPVGYEINNVVQLQDPVGNTFNLSYYFNQGDHRIGAGLFNILSVHEIQHTVMVH 430
Query: 70 FEYKDTSHFGVHIFDKSTLEIRY 92
F Y+ + F +HIFD + EIRY
Sbjct: 431 FLYRGQARFNIHIFDPFSREIRY 499
>TC18004
Length = 713
Score = 43.5 bits (101), Expect = 7e-05
Identities = 17/24 (70%), Positives = 21/24 (86%)
Frame = +2
Query: 434 GDACVFELINKEDPVLDVHIYRGH 457
GD CVFELI++ED V DVHI++GH
Sbjct: 299 GDVCVFELIHQEDAVFDVHIFKGH 370
>TC10273
Length = 520
Score = 35.0 bits (79), Expect = 0.026
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 7/66 (10%)
Frame = +2
Query: 214 HIPSFFAKKYLKNETDVLLQVLDGRTWSVSF-------NLGKFNAGWKKFTSVNNLKVGD 266
H+P++ +T+++L+ G W+V+ + F GW F N++ GD
Sbjct: 23 HLPAY--------KTEIMLRNSRGECWTVNSVPDAKGRTVHTFCGGWXAFVRDNDINFGD 178
Query: 267 VCLFEL 272
C+FEL
Sbjct: 179 TCIFEL 196
Score = 29.6 bits (65), Expect = 1.1
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = +2
Query: 421 GWVEFAKKSKLEVGDACVFELI 442
GW F + + + GD C+FEL+
Sbjct: 134 GWXAFVRDNDINFGDTCIFELV 199
>TC16790
Length = 584
Score = 32.3 bits (72), Expect = 0.17
Identities = 12/40 (30%), Positives = 22/40 (55%)
Frame = +2
Query: 417 RLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYRG 456
R+ GW +F ++ + GD C F+ N + +++V I G
Sbjct: 302 RIGSGWKKFCTRNGVRAGDLCCFKFTNIAERIVEVSIVFG 421
Score = 32.0 bits (71), Expect = 0.22
Identities = 12/37 (32%), Positives = 21/37 (56%)
Frame = +2
Query: 248 KFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLI 284
+ +GWKKF + N ++ GD+C F+ ++V I
Sbjct: 302 RIGSGWKKFCTRNGVRAGDLCCFKFTNIAERIVEVSI 412
>TC14104 GB|CAA61590.1|897773|LJAS2GENE asparagine synthase
(glutamine-hydrolysing) {Lotus corniculatus var.
japonicus;} , complete
Length = 2259
Score = 30.8 bits (68), Expect = 0.49
Identities = 15/50 (30%), Positives = 23/50 (46%)
Frame = -2
Query: 330 KRTSKLISPCPLKNAALEEANKFNSNNPFFIVNIAPDKNRDYRPRVPKLF 379
K + L+ P P++ L+ A K NPF + I K+ Y K+F
Sbjct: 287 KVITMLVEPTPVRATVLQSARKLKDANPFGLRVIRTTKHSKYATHCDKIF 138
>TC15236 homologue to UP|GCST_PEA (P49364) Aminomethyltransferase,
mitochondrial precursor (Glycine cleavage system T
protein) (GCVT) , complete
Length = 1457
Score = 29.3 bits (64), Expect = 1.4
Identities = 13/34 (38%), Positives = 22/34 (64%), Gaps = 2/34 (5%)
Frame = -3
Query: 253 WKKFTSVNNLKVGDVCLFELN--KSVPLSLKVLI 284
W S+NN++V + CLF+L+ + +P S +LI
Sbjct: 240 WHHLPSINNMEVIESCLFQLSL*RKIPTSNSLLI 139
>TC13356 weakly similar to UP|Q9SVG3 (Q9SVG3) Reticuline oxidase-like
protein, partial (16%)
Length = 433
Score = 28.9 bits (63), Expect = 1.8
Identities = 12/27 (44%), Positives = 14/27 (51%)
Frame = +1
Query: 118 IPPCKKTKLKSPMSCPKPRKKLRTSTS 144
+PPCK K P S PKP K S +
Sbjct: 280 LPPCKNLTSKPPSSAPKPLKSSSKSAA 360
>BP050885
Length = 559
Score = 28.1 bits (61), Expect = 3.2
Identities = 16/33 (48%), Positives = 19/33 (57%)
Frame = -1
Query: 412 KKGPSRLSKGWVEFAKKSKLEVGDACVFELINK 444
KKG LS GW FA L GDA +F+L N+
Sbjct: 430 KKG---LSGGWRGFAIAHDLADGDALIFQLTNR 341
>BP029246
Length = 455
Score = 28.1 bits (61), Expect = 3.2
Identities = 18/46 (39%), Positives = 22/46 (47%), Gaps = 2/46 (4%)
Frame = -3
Query: 247 GKFNAGWKKFTSVNNLKVGDVCLFELNKSV--PLSLKVLIFPLAEE 290
G GWK F NNL+ DVC+ V L + V IF + EE
Sbjct: 378 GGLTNGWKHFAVDNNLEEFDVCVLTPAGKVNNTLIVDVSIFRVVEE 241
Score = 27.7 bits (60), Expect = 4.1
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Frame = -3
Query: 418 LSKGWVEFAKKSKLEVGDACVFELINKEDP--VLDVHIYR 455
L+ GW FA + LE D CV K + ++DV I+R
Sbjct: 372 LTNGWKHFAVDNNLEEFDVCVLTPAGKVNNTLIVDVSIFR 253
>AV409011
Length = 436
Score = 28.1 bits (61), Expect = 3.2
Identities = 12/41 (29%), Positives = 19/41 (46%)
Frame = -2
Query: 135 PRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEG 175
P K+ T + ELHN P + + ++ TT C +G
Sbjct: 354 PLPKISLLTRVQLNSLVELHNRPGNITLSEEGHSTTNCKKG 232
>BP061842
Length = 318
Score = 27.7 bits (60), Expect = 4.1
Identities = 19/65 (29%), Positives = 27/65 (41%)
Frame = +3
Query: 241 SVSFNLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEEPHSLPRSLVQ 300
S LG + W + TS+N N+S+ L ++ EEP RSLV
Sbjct: 111 SAKQQLGSHHVHWPQTTSINR-----------NQSISLEAQMQSLHSIEEPAGRKRSLVA 257
Query: 301 GDRVN 305
G +N
Sbjct: 258 GWHLN 272
>TC10231 similar to UP|O23110 (O23110) AP2 domain containing protein RAP2.8
(Fragment), partial (22%)
Length = 707
Score = 27.3 bits (59), Expect = 5.4
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = +1
Query: 418 LSKGWVEFAKKSKLEVGDACVF 439
L+KGW F K+ L GDA F
Sbjct: 130 LTKGWSRFVKEKNLRAGDAVRF 195
>TC7891 similar to UP|DCAM_VICFA (Q9M4D8) S-adenosylmethionine decarboxylase
proenzyme (AdoMetDC) (SamDC) [Contains:
S-adenosylmethionine decarboxylase alpha chain;
S-adenosylmethionine decarboxylase beta chain] , complete
Length = 1889
Score = 27.3 bits (59), Expect = 5.4
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = +1
Query: 266 DVCLFELNKSVPLSLKVLIF 285
D CLFELNKS+P ++ F
Sbjct: 1624 DFCLFELNKSLPSGFGIVPF 1683
>AV770380
Length = 526
Score = 27.3 bits (59), Expect = 5.4
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = -1
Query: 435 DACVFELINKEDPVLDVHIYR 455
D C FELI K + VHI+R
Sbjct: 526 DVCTFELIAKTNNTFQVHIFR 464
>AU089318
Length = 641
Score = 26.9 bits (58), Expect = 7.0
Identities = 15/41 (36%), Positives = 23/41 (55%)
Frame = +3
Query: 233 QVLDGRTWSVSFNLGKFNAGWKKFTSVNNLKVGDVCLFELN 273
+V D W++ LG N +KK+ + N +VGD +FE N
Sbjct: 111 KVGDSAGWTI---LG--NVDYKKWAAPXNFQVGDTIIFEYN 218
>TC11614 similar to UP|Q9FHH4 (Q9FHH4) Diacylglycerol kinase-like protein,
partial (24%)
Length = 634
Score = 26.6 bits (57), Expect = 9.2
Identities = 11/34 (32%), Positives = 17/34 (49%)
Frame = +3
Query: 33 GTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGH 66
G WK + + DG+ W Q KDF+ + + H
Sbjct: 252 GGGWKNAYMQMDGEPWKQPLSKDFSTFVEIKREH 353
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,659,356
Number of Sequences: 28460
Number of extensions: 128090
Number of successful extensions: 639
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 628
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 638
length of query: 457
length of database: 4,897,600
effective HSP length: 93
effective length of query: 364
effective length of database: 2,250,820
effective search space: 819298480
effective search space used: 819298480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0040.4


