

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.4
(616 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
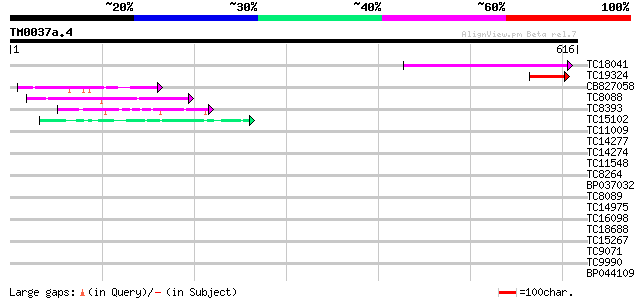
Score E
Sequences producing significant alignments: (bits) Value
TC18041 weakly similar to UP|O80588 (O80588) At2g44190 protein, ... 100 7e-22
TC19324 UP|Q84P50 (Q84P50) Elicitin-like protein (Fragment), par... 79 2e-15
CB827058 63 2e-10
TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like arabinogalac... 44 8e-05
TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenyla... 43 2e-04
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 40 8e-04
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 40 0.001
TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_1... 40 0.001
TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.3... 39 0.002
TC11548 similar to UP|Q9FG37 (Q9FG37) Gamma-tubulin interacting ... 39 0.003
TC8264 similar to GB|AAK20861.1|13377784|AF333974 fasciclin-like... 38 0.004
BP037032 38 0.005
TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinog... 38 0.005
TC14975 homologue to UP|Q8LG37 (Q8LG37) AtPH1-like protein, part... 38 0.005
TC16098 homologue to UP|Q8LAQ5 (Q8LAQ5) Flower pigmentation prot... 37 0.007
TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%) 37 0.007
TC15267 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Ce... 37 0.009
TC9071 similar to UP|Q9LT85 (Q9LT85) Similarity to receptor prot... 37 0.009
TC9990 similar to UP|Q9FJW0 (Q9FJW0) RuvB DNA helicase-like prot... 37 0.009
BP044109 37 0.009
>TC18041 weakly similar to UP|O80588 (O80588) At2g44190 protein, partial
(11%)
Length = 925
Score = 100 bits (249), Expect = 7e-22
Identities = 61/183 (33%), Positives = 104/183 (56%)
Frame = +3
Query: 429 MQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQQLKLELKLNSVLN 488
+QWRFANA+A A + Q +K LY+ + + +SV RK+ L+ L+ L+++L+
Sbjct: 3 LQWRFANAKAAASMKAQQKESEKALYSHAMKISEMRDSVNRKRAELELLQRYKTLSTILD 182
Query: 489 DQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVDIEHLKVAICQAVDVMQA 548
QM YL+EW+++E D+ +L+ A + L ++++LPV VD + A+ A +M+
Sbjct: 183 AQMPYLEEWSSMEEDYSVSLTEATQALVNASVQLPVGGNVRVDEREVGEALNSASKMMET 362
Query: 549 MGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVAALQVEESSLQTHLIQF 608
+ S I+ L + E + IS +A V E++++ E LL+ QVEE SL+ LIQ
Sbjct: 363 IVSNIQRLVPKAEETDSSISELARVVGGERALIGESGDLLSKTYKSQVEECSLRGQLIQL 542
Query: 609 KQV 611
+
Sbjct: 543 HSI 551
>TC19324 UP|Q84P50 (Q84P50) Elicitin-like protein (Fragment), partial (6%)
Length = 505
Score = 79.0 bits (193), Expect = 2e-15
Identities = 42/44 (95%), Positives = 42/44 (95%)
Frame = +2
Query: 565 DLISGVAVVATKEKSILDECEMLLASVAALQVEESSLQTHLIQF 608
DLISGVAVVATK KSI DECEMLLASVAALQVEESSLQTHLIQF
Sbjct: 2 DLISGVAVVATKAKSIPDECEMLLASVAALQVEESSLQTHLIQF 133
>CB827058
Length = 555
Score = 62.8 bits (151), Expect = 2e-10
Identities = 64/172 (37%), Positives = 80/172 (46%), Gaps = 14/172 (8%)
Frame = +1
Query: 9 ASSRRLKAVETP-RPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGS---- 63
A + K TP RPPL+ N A +RR ++REVTSRY S + S+ S S
Sbjct: 73 AETTTTKRAPTPTRPPLL--PSGNVAPPPPLSRRVKSREVTSRYMSSSSSSSSSSSVSSP 246
Query: 64 -RRCPSPNLTRTATPA---ASLQLLP----KRSLSAERKRPSTPPSPPSRRLSTPVNDSA 115
RR SP + RT + +S +P KRS SAER+R P R V SA
Sbjct: 247 PRRYHSPIVPRTVNSSKQKSSTIAVPTPQVKRSQSAERQRQRPRQGTP-RPSGAGVVASA 423
Query: 116 IDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSK-KERPVTS 166
D P H + ++ RSLSVSFQ + SI VSK K PV S
Sbjct: 424 ADA------------PAAHKMLFTSTRSLSVSFQGESFSIQVSKPKPAPVQS 543
>TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (69%)
Length = 1527
Score = 43.9 bits (102), Expect = 8e-05
Identities = 46/188 (24%), Positives = 78/188 (41%), Gaps = 7/188 (3%)
Frame = +2
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPN-LTRTATP 77
+P P + + T+ AR S T + + S TPP PS + PSP T + +
Sbjct: 164 SPNTPTSPPSTTTSPSPTSPAR-STTAKPSPSSPSTTPPCPPSSTSTSPSPP*KTSSPST 340
Query: 78 AASLQLLPKRSLSAERKRPST-----PPSPPSRRLSTPVNDSAIDGR-LSSRKVAASRLP 131
++S L PK S + PS+ PP+PP +T + ++ + + S + +
Sbjct: 341 SSSTTLAPKSSTR*PTEPPSSPPCSRPPAPPPEPPATSTSPTSKEAK*ASPLRTTTASTH 520
Query: 132 EGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRK 191
N W S + S+ +S P + K RP A + N A + + S RK
Sbjct: 521 STSNQWKS-FHTSSLFSKSLNP*APPTPKHRPQLRAISTSSPSCPNKAAKPSQIYSEPRK 697
Query: 192 ATPERKRS 199
P +++
Sbjct: 698 LFPHSRKT 721
Score = 28.9 bits (63), Expect = 2.5
Identities = 22/76 (28%), Positives = 33/76 (42%), Gaps = 1/76 (1%)
Frame = +2
Query: 321 PHTDRAGLSIAGVRSQSLSAPGSRLPSPSRIS-VQSTSSSRGVSPSRSRPSTPPSRGGVS 379
P A ++ + ++P S SPS S +ST++ S + P PPS S
Sbjct: 125 PPPPMAATTLPAFSPNTPTSPPSTTTSPSPTSPARSTTAKPSPSSPSTTPPCPPSSTSTS 304
Query: 380 PSRIRPTSSSIQSNDS 395
PS TSS S+ +
Sbjct: 305 PSPP*KTSSPSTSSST 352
>TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenylated Rab
receptor 2 {Arabidopsis thaliana;}, partial (78%)
Length = 1012
Score = 42.7 bits (99), Expect = 2e-04
Identities = 59/202 (29%), Positives = 83/202 (40%), Gaps = 33/202 (16%)
Frame = +1
Query: 53 SPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSP---------- 102
+P PP PS SPN + TA SL +P RSL P+ PSP
Sbjct: 133 TPLPPPPPSAPSSTTSPNPSATA----SLSAVPGRSLPIAPHSPNRNPSPMPPSASARIS 300
Query: 103 --------PSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIIS 154
PS R S+P S I R S A LP GH+ S +R +++SF S S
Sbjct: 301 PTSASTTTPSFRSSSPCRSSPIRSR--SSP*PAFSLP-GHSSTSSALR-IALSFSSAAPS 468
Query: 155 IPVSKKER------------PVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLK 202
P+S++ R PV+ S L +++ +++R + RSPL+
Sbjct: 469 -PISRRSRFSPH*RSSSFSSPVSVPSSSLLSCSASPLFASMAHSASLR-ISSLMSRSPLR 642
Query: 203 GKNASDQSE---NSKPVDSLPS 221
++S S S P LPS
Sbjct: 643 PPDSSPSSALLLPSPPPRFLPS 708
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 40.4 bits (93), Expect = 8e-04
Identities = 60/234 (25%), Positives = 88/234 (36%)
Frame = +1
Query: 33 ATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAE 92
ATA+ ++ T TS K+ T S P +R+ TP +S P L+
Sbjct: 145 ATASPSSEPCTTATTTSTTKAATNSSPPE----------SRSTTPPSSE---PTCLLA-- 279
Query: 93 RKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDI 152
P++PP+P S STP ++S P+ N T S S
Sbjct: 280 ---PASPPTPKSSHSSTPPPSR-----------SSSTTPKSKNATSLTAPSCPPP-ASPA 414
Query: 153 ISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNASDQSEN 212
+ P K P + + +P+S + TPS++ A P SP+ ++ + E
Sbjct: 415 ATAPAPSKTPP--NPPTNSSKPSSCKSSLPNTTPSSLSSAPPSPTTSPISTPSSPENQE- 585
Query: 213 SKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSL 266
KP + PS P R SSS +N A K PAS S L
Sbjct: 586 -KPASTPPSA-------PPRSTSSSSSSPINTPRKPSSA--TKAPASSPPGSEL 717
Score = 35.4 bits (80), Expect = 0.027
Identities = 39/149 (26%), Positives = 57/149 (38%), Gaps = 6/149 (4%)
Frame = +1
Query: 5 ESQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSG-S 63
+S+ A+S + P P A + S+ S KS P +TPS S
Sbjct: 355 KSKNATSLTAPSCPPPASPAATAPAPSKTPPNPPTNSSKP----SSCKSSLPNTTPSSLS 522
Query: 64 RRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSR 123
PSP + +TP++ P+ PS PP S S+P+N + SS
Sbjct: 523 SAPPSPTTSPISTPSS-----PENQEKPASTPPSAPPRSTSSSSSSPINTPR---KPSSA 678
Query: 124 KVAASRLPEG-----HNLWPSTMRSLSVS 147
A + P G H+ PS R S +
Sbjct: 679 TKAPASSPPGSELSLHHSPPSASRKSSTT 765
Score = 28.1 bits (61), Expect = 4.3
Identities = 23/77 (29%), Positives = 32/77 (40%)
Frame = +1
Query: 317 RSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRG 376
RS+TP + +A + + S P PSR S ST+ + S + PS PP
Sbjct: 235 RSTTPPSSEPTCLLAPASPPTPKSSHSSTPPPSRSS--STTPKSKNATSLTAPSCPPP-- 402
Query: 377 GVSPSRIRPTSSSIQSN 393
SP+ P S N
Sbjct: 403 -ASPAATAPAPSKTPPN 450
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 40.0 bits (92), Expect = 0.001
Identities = 47/152 (30%), Positives = 57/152 (36%), Gaps = 14/152 (9%)
Frame = -2
Query: 268 RLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDRAG 327
RL +S P +SS L L G+ S SL S +
Sbjct: 450 RLEGLSSSSPPSSESSSPEPSLSDLFLFD--GTPAFSPSSLSLSSPSSSSGSKSLDASSS 277
Query: 328 LSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRP-STPPSR----------- 375
+ A VR S P S S S S S+SSS SP RP STPPS
Sbjct: 276 SAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSP 97
Query: 376 --GGVSPSRIRPTSSSIQSNDSVSVLSFIADF 405
G S S P+SSS S+ S S L + +F
Sbjct: 96 PPGASSSSSSSPSSSSPSSSSSSSPLILVREF 1
Score = 38.9 bits (89), Expect = 0.002
Identities = 51/169 (30%), Positives = 66/169 (38%), Gaps = 2/169 (1%)
Frame = -2
Query: 226 QHRW--PSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSLRRLSLSEEASKPLQRAS 283
+HRW P R+ G SSS P+S SS SLS+ A
Sbjct: 474 KHRWIQPYRLEGLSSSSP---------------PSSE---SSSPEPSLSDLFLFDGTPAF 349
Query: 284 SDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGS 343
S S LS +S + + + +R R S P + + S + S S S+ GS
Sbjct: 348 SPSSLSLSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGS 169
Query: 344 RLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQS 392
P P S +SSS S S S P S SPS P+SSS S
Sbjct: 168 P-PPPRPPSTPPSSSSSSSSSSSSPPPGASSSSSSSPSSSSPSSSSSSS 25
Score = 30.0 bits (66), Expect = 1.1
Identities = 23/77 (29%), Positives = 35/77 (44%), Gaps = 7/77 (9%)
Frame = -2
Query: 53 SPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPS-------PPSR 105
S + PS+ SGS+ + + + AT + +PK S S+ S+ S PP R
Sbjct: 333 SLSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPR 154
Query: 106 RLSTPVNDSAIDGRLSS 122
STP + S+ SS
Sbjct: 153 PPSTPPSSSSSSSSSSS 103
Score = 27.3 bits (59), Expect = 7.4
Identities = 23/106 (21%), Positives = 47/106 (43%)
Frame = -2
Query: 6 SQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRR 65
S+ + A T R K++++++++++ S + + P PPSTP S
Sbjct: 303 SKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSS-- 130
Query: 66 CPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPV 111
+++ ++S P + S+ PS+ SP S S+P+
Sbjct: 129 --------SSSSSSSSSSPPPGASSSSSSSPSS-SSPSSSSSSSPL 19
>TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_13), partial
(33%)
Length = 941
Score = 39.7 bits (91), Expect = 0.001
Identities = 27/84 (32%), Positives = 42/84 (49%)
Frame = +3
Query: 22 PPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASL 81
PPL+ + TA T+ SR ++ + +PP P S SP +T TA P ++
Sbjct: 351 PPLLTTTSPPSVTAPRTSSASRLPSSSALDAALSPP-LPCTSSGPSSPPVTTTAPPCTAI 527
Query: 82 QLLPKRSLSAERKRPSTPPSPPSR 105
+ +RS + +RP+T SPP R
Sbjct: 528 SPMTRRSTAP--RRPATSRSPPLR 593
Score = 32.0 bits (71), Expect = 0.30
Identities = 36/169 (21%), Positives = 60/169 (35%), Gaps = 5/169 (2%)
Frame = +3
Query: 44 TREVTSRYKSPTPPSTP----SGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTP 99
+R S SP+PP P + S P+P+ T TP+ P +
Sbjct: 90 SRSPPSSSSSPSPPPPPHAPAAASSSPPTPSATLPPTPSP--------------PSPRSD 227
Query: 100 PSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSK 159
PSP S + P S+ + ++ K + H + L+ + + + S
Sbjct: 228 PSPLSTSTTNPTTSSSTTPKTTTNKSNPNTARLSHAAP*GSPPLLTTTSPPSVTAPRTSS 407
Query: 160 KER-PVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNAS 207
R P +SA D L P +P A P SP+ ++ +
Sbjct: 408 ASRLPSSSALDAALSPPLPCTSSGPSSPPVTTTAPPCTAISPMTRRSTA 554
>TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.35
{Arabidopsis thaliana;} , partial (16%)
Length = 1208
Score = 38.9 bits (89), Expect = 0.002
Identities = 33/108 (30%), Positives = 46/108 (42%), Gaps = 2/108 (1%)
Frame = +3
Query: 11 SRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPST--PSGSRRCPS 68
SR + + P + AT++ T+ T S +PT P+T PSG+ PS
Sbjct: 66 SRWISTSPSSPPSISRTSTGKPATSSRTSSSGSTPTAVSP-PNPTTPATPPPSGTSVSPS 242
Query: 69 PNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAI 116
+LT + TP + P RS + P P SPPS S S I
Sbjct: 243 LSLTPSLTPFS-----PSRSSTPNPPIPPNPSSPPSASSSPTSTTSMI 371
Score = 26.9 bits (58), Expect = 9.7
Identities = 23/60 (38%), Positives = 30/60 (49%), Gaps = 12/60 (20%)
Frame = +3
Query: 334 RSQSLSAPGSRLPSPSRISV-----QSTSSSRGVSPSR------SRPSTPPSRG-GVSPS 381
RS+ +S S PS SR S S +SS G +P+ + P+TPP G VSPS
Sbjct: 63 RSRWISTSPSSPPSISRTSTGKPATSSRTSSSGSTPTAVSPPNPTTPATPPPSGTSVSPS 242
>TC11548 similar to UP|Q9FG37 (Q9FG37) Gamma-tubulin interacting
protein-like, partial (6%)
Length = 480
Score = 38.5 bits (88), Expect = 0.003
Identities = 40/137 (29%), Positives = 55/137 (39%), Gaps = 6/137 (4%)
Frame = +3
Query: 36 TATARRSRTREVTSRYKSP------TPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSL 89
T T R + T SP +PP TP PSPN + +P L P
Sbjct: 96 TLTPRNFKNPSATPSASSPAASTPLSPPPTPP-----PSPNKSNAVSP---LTAAPPTHS 251
Query: 90 SAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQ 149
+ PS+PP PP + P + S+ SS+K+A H L PS S+ SF
Sbjct: 252 RSPTYTPSSPPEPPLLTTNGPCSTSS----RSSQKIAR------HWLRPS---SIPPSFF 392
Query: 150 SDIISIPVSKKERPVTS 166
S+P + P T+
Sbjct: 393 PPSQSLPTTTTRDPTTA 443
>TC8264 similar to GB|AAK20861.1|13377784|AF333974 fasciclin-like
arabinogalactan-protein 9 {Arabidopsis thaliana;},
partial (38%)
Length = 942
Score = 38.1 bits (87), Expect = 0.004
Identities = 40/149 (26%), Positives = 63/149 (41%), Gaps = 14/149 (9%)
Frame = +1
Query: 6 SQQASSRRLKAVETPRPPLVLA---------EKNNAATATATARRSRTREVTSRYKSPTP 56
S + S LK+ PR L A + N ++A++R +P P
Sbjct: 52 SHSSPSSLLKSKPKPRHRLPPAR*T*PQSSKKPGNTPPSSASSRNPNNSPKLRANSTPPP 231
Query: 57 PSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTP-----V 111
++PS S R +P+ T + P+ + + K S + R ST PSP S++ +TP +
Sbjct: 232 KASPS-SPRPTTPSKTSSLAPSMTSPMTRK*SSFSTMSRQSTTPSPISKQ*ATP*GHRLL 408
Query: 112 NDSAIDGRLSSRKVAASRLPEGHNLWPST 140
A+ G S K P G +L ST
Sbjct: 409 RKKALGGLTSRVKGTK*M*PPGWSLLLST 495
Score = 28.9 bits (63), Expect = 2.5
Identities = 23/65 (35%), Positives = 32/65 (48%)
Frame = +1
Query: 336 QSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSNDS 395
QS PG+ PS ++S + SP STPP + SPS RPT+ S S+ +
Sbjct: 133 QSSKKPGNTPPS------SASSRNPNNSPKLRANSTPPPKA--SPSSPRPTTPSKTSSLA 288
Query: 396 VSVLS 400
S+ S
Sbjct: 289 PSMTS 303
Score = 28.5 bits (62), Expect = 3.3
Identities = 19/60 (31%), Positives = 30/60 (49%), Gaps = 1/60 (1%)
Frame = +1
Query: 339 SAPGSRLPSPS-RISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSNDSVS 397
S+ SR P+ S ++ ST + SPS RP+TP ++PS P + S ++S
Sbjct: 166 SSASSRNPNNSPKLRANSTPPPKA-SPSSPRPTTPSKTSSLAPSMTSPMTRK*SSFSTMS 342
>BP037032
Length = 577
Score = 37.7 bits (86), Expect = 0.005
Identities = 43/151 (28%), Positives = 59/151 (38%), Gaps = 20/151 (13%)
Frame = +3
Query: 13 RLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLT 72
R A R P A AA +T +S +R T+ S +PPS+P CP P+
Sbjct: 78 REPAATEKRAPSSPATCLYAAHPASTPDKSSSRRRTA---SSSPPSSP*----CPRPHRL 236
Query: 73 RTAT---------------PAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAID 117
R+ T P + P+ L A P+ PSPPSRR + +
Sbjct: 237 RSETSRVGTHPDPPTCPDPPTTTPCKTPRSRL*ARCSHPA--PSPPSRRAIARSGSAGVA 410
Query: 118 GRLSSR-----KVAASRLPEGHNLWPSTMRS 143
G SR + +LP L+P T RS
Sbjct: 411 GMSPSRWR*GKAFSRHQLPSHRRLFPRTRRS 503
>TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (43%)
Length = 890
Score = 37.7 bits (86), Expect = 0.005
Identities = 38/150 (25%), Positives = 53/150 (35%)
Frame = +2
Query: 48 TSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRL 107
TS SP+ PS+P + PSP +T P + +T SPPSR
Sbjct: 233 TSPAFSPSTPSSPPSTTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTSSQNTHQSPPSRTS 412
Query: 108 STPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSA 167
S + S SS +R P P + P + P +S
Sbjct: 413 SPSTSSSTTSAPRSS-----TRSPTAPPSPPQCTKP------------PAPPRAPPDSST 541
Query: 168 SDRTLRPASNVAHRQGETPSTIRKATPERK 197
S + S+ A R PS +R + P RK
Sbjct: 542 SPTSAAGKSDSAPRTTTAPSPLRSSNP*RK 631
Score = 33.5 bits (75), Expect = 0.10
Identities = 42/178 (23%), Positives = 63/178 (34%), Gaps = 7/178 (3%)
Frame = +2
Query: 48 TSRYKSPTPPSTPSGSRRCPSP-NLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRR 106
T SPT S S P L+R ++ L P+ ++ PSTP SPP
Sbjct: 104 THHTSSPTQISPRQCSCSARQPWRLSRQRCSSSPWLLPPQTPTTSPAFSPSTPSSPP--- 274
Query: 107 LSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTS 166
ST + S S+ + P W ++ ++ S P S+ P TS
Sbjct: 275 -STTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTSSQNTHQS--------PPSRTSSPSTS 427
Query: 167 AS------DRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNASDQSENSKPVDS 218
+S T P + + Q P +A P+ SP SD + + S
Sbjct: 428 SSTTSAPRSSTRSPTAPPSPPQCTKPPAPPRAPPDSSTSPTSAAGKSDSAPRTTTAPS 601
Score = 33.1 bits (74), Expect = 0.13
Identities = 41/159 (25%), Positives = 60/159 (36%), Gaps = 12/159 (7%)
Frame = +2
Query: 22 PPLVLAEKNN-----AATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTAT 76
PP + +N + T++ + S T S +SPT P +P +C P A
Sbjct: 356 PPWTTSSQNTHQSPPSRTSSPSTSSSTTSAPRSSTRSPTAPPSPP---QCTKPPAPPRAP 526
Query: 77 PAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNL 136
P +S S R +T PSP R S P S SSR V P+ L
Sbjct: 527 PDSSTSPTSAAGKSDSAPRTTTAPSP--LRSSNP*RKSPTTSP-SSRSVRFFPPPQQKPL 697
Query: 137 WPSTMRSLS-------VSFQSDIISIPVSKKERPVTSAS 168
P + +S V+ S +S P + +P + S
Sbjct: 698 HPHPLSRISPPLCPSTVARSSPTLSPPRRTRTQPSPTTS 814
Score = 27.7 bits (60), Expect = 5.7
Identities = 31/93 (33%), Positives = 37/93 (39%), Gaps = 21/93 (22%)
Frame = +2
Query: 318 SSTPHTDRAGLSIAGVRS----QSLSAPGSRLP-----------SPSRISVQSTSSSRGV 362
SS P T + + RS QS SAP + P PSR S STSSS
Sbjct: 263 SSPPSTTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTSSQNTHQSPPSRTSSPSTSSSTTS 442
Query: 363 SPSRSR------PSTPPSRGGVSPSRIRPTSSS 389
+P S PS P +P R P SS+
Sbjct: 443 APRSSTRSPTAPPSPPQCTKPPAPPRAPPDSST 541
>TC14975 homologue to UP|Q8LG37 (Q8LG37) AtPH1-like protein, partial (86%)
Length = 892
Score = 37.7 bits (86), Expect = 0.005
Identities = 27/106 (25%), Positives = 45/106 (41%)
Frame = +3
Query: 6 SQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRR 65
S+Q ++ L + TP P K + + A A +E +S K+P ++PS +
Sbjct: 132 SRQPTTTALNSGPTPNAPAGSQNKASTSKPGAAAGSFSNKENSSGSKTPPSRASPSHAAS 311
Query: 66 CPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPV 111
PSP ++ + P S+ PS P P R S+P+
Sbjct: 312 SPSPPVSPSRAP----------KTSSTNPTPSNSPRAPIRCTSSPI 419
Score = 33.9 bits (76), Expect = 0.079
Identities = 47/181 (25%), Positives = 69/181 (37%), Gaps = 18/181 (9%)
Frame = +3
Query: 32 AATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSA 91
A A ARRSR T+ PT P+ P+GS+ N T+ P A+ + S+
Sbjct: 102 ACGARRPARRSRQPTTTALNSGPT-PNAPAGSQ-----NKASTSKPGAAAGSFSNKENSS 263
Query: 92 ERKRPSTPPS--PPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQ 149
K TPPS PS S+P S V+ SR P+ + P+ S +
Sbjct: 264 GSK---TPPSRASPSHAASSP-----------SPPVSPSRAPKTSSTNPTPSNSPRAPIR 401
Query: 150 SDIISIPVSKKE----------------RPVTSASDRTLRPASNVAHRQGETPSTIRKAT 193
I + ++ P+ +S T A +A R+ ST R+A
Sbjct: 402 CTSSPILIRRRRTGSTLLAAPSFSTPDPSPILRSSIMTAPNADGIAPRRSR--STRRRAA 575
Query: 194 P 194
P
Sbjct: 576 P 578
>TC16098 homologue to UP|Q8LAQ5 (Q8LAQ5) Flower pigmentation protein ATAN11,
partial (79%)
Length = 919
Score = 37.4 bits (85), Expect = 0.007
Identities = 37/124 (29%), Positives = 49/124 (38%), Gaps = 5/124 (4%)
Frame = +1
Query: 20 PRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAA 79
PRP L + NA T++ S + P PP P S S TRTA+ A
Sbjct: 313 PRPFSSLTKSVNALTSSPPPATSSACGGSPTPMKPNPPPPPPASNSRASLTETRTASIVA 492
Query: 80 --SLQLLPKRSLSAERKRPSTPPSP---PSRRLSTPVNDSAIDGRLSSRKVAASRLPEGH 134
+ K SL+A STPP+P R+ S P + I +SR A P
Sbjct: 493 R*PPSIGTKLSLAASEPPASTPPAPFGISRRKPSIPSSSRMIRRFTTSRGAALGFSPPSP 672
Query: 135 NLWP 138
+ P
Sbjct: 673 PMAP 684
Score = 30.0 bits (66), Expect = 1.1
Identities = 27/80 (33%), Positives = 38/80 (46%), Gaps = 5/80 (6%)
Frame = +1
Query: 314 RPHRSSTPHTDRAGLSIAGVRSQSLSAPGS-RLPSPSRISVQSTSSSRG----VSPSRSR 368
+PH +STP T G S A + S S P S P+ SR S +T ++R +SPS +
Sbjct: 133 KPHGTSTP*T---GASAATKSTASPSPPSSNNTPTASRSSSSTTPTARSDPTRISPSNTH 303
Query: 369 PSTPPSRGGVSPSRIRPTSS 388
P ++ S TSS
Sbjct: 304 THPPRPFSSLTKSVNALTSS 363
>TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%)
Length = 514
Score = 37.4 bits (85), Expect = 0.007
Identities = 39/128 (30%), Positives = 54/128 (41%)
Frame = +3
Query: 13 RLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLT 72
R P PP +A+TAT T S SR SP PS P+ S SP
Sbjct: 180 RFSGTPPPPPPAT----GSASTATPTPPTS------SRSASPPSPS-PANSHMASSP--- 317
Query: 73 RTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPE 132
+P ++L + S+S S P SPP+ R +T + S I +SSR++
Sbjct: 318 --PSPTSALSV----SVSTPSPAHSPPTSPPAPRSATSTS-SRISSPVSSRRLFHVSPAL 476
Query: 133 GHNLWPST 140
+ WP T
Sbjct: 477 SASTWPPT 500
>TC15267 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein), partial (6%)
Length = 1003
Score = 37.0 bits (84), Expect = 0.009
Identities = 30/102 (29%), Positives = 44/102 (42%), Gaps = 11/102 (10%)
Frame = +1
Query: 13 RLKAVETPRPPLVLAEKNNAATATATAR-------RSRTREVTSRYKSPTPPST----PS 61
R K T ++ NN A T T + S + S + SP+PPS+ P
Sbjct: 52 RKKNTTTTTTTTTTSKTNNNAINTTTVKLPPP*SPSSPSSSSLSPWLSPSPPSSTTPPPP 231
Query: 62 GSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPP 103
S R + NL+ TP+A + S + P++PPSPP
Sbjct: 232 PSHRTNNNNLSTQPTPSA------PSATSLASRTPASPPSPP 339
Score = 31.6 bits (70), Expect = 0.39
Identities = 46/156 (29%), Positives = 58/156 (36%), Gaps = 17/156 (10%)
Frame = +1
Query: 29 KNNAATATATARRSRTREV---TSRYKSPTP--PSTPSGSRRCPSPNLTRTATPAASLQL 83
K N T T T S+T T+ K P P PS+PS S P
Sbjct: 55 KKNTTTTTTTTTTSKTNNNAINTTTVKLPPP*SPSSPSSSSLSPW--------------- 189
Query: 84 LPKRSLSAERKRPSTPPSPPSRR-----LSTPVNDSAIDG-RLSSRKVAA------SRLP 131
LS +TPP PPS R LST SA L+SR A+ + P
Sbjct: 190 -----LSPSPPSSTTPPPPPSHRTNNNNLSTQPTPSAPSATSLASRTPASPPSPPPTTTP 354
Query: 132 EGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSA 167
HN PS+ + + S S P S+ V +A
Sbjct: 355 PTHN--PSSPSPSAPPWTSSPPSPPRSESRPQVLTA 456
Score = 27.3 bits (59), Expect = 7.4
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Frame = +1
Query: 343 SRLPSPSRISVQSTSSSRGVSPSRSRP-STPPSRGGVSPSRIRPTSSS 389
S P+PS S S +S SP P +TPP+ SPS P +S
Sbjct: 262 STQPTPSAPSATSLASRTPASPPSPPPTTTPPTHNPSSPSPSAPPWTS 405
>TC9071 similar to UP|Q9LT85 (Q9LT85) Similarity to receptor protein
kinase, partial (15%)
Length = 872
Score = 37.0 bits (84), Expect = 0.009
Identities = 51/178 (28%), Positives = 70/178 (38%), Gaps = 7/178 (3%)
Frame = +2
Query: 20 PRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAA 79
P P L N++A T A S TS TP +TP P+P TA+ AA
Sbjct: 275 PPPAAHLPHPNHSAV*TKLAASSSNSPPTST----TPTATPKPGTAKPTP-ANSTASDAA 439
Query: 80 SLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPS 139
+Q P R S E STPP+ P++ + S+ ASR P+
Sbjct: 440 YIQT-PTREPSPEST--STPPASPAKTATLSP---------STASSTASRNSHSST*TPT 583
Query: 140 TMRSLSVSFQSDIISIPVSKKERPVTSASDRTLR-------PASNVAHRQGETPSTIR 190
T +LS + S S P S T+ S + R P+S + G T +T R
Sbjct: 584 TSPALSRAKSS---SSPTSTSSTSATTNSPASFRWMFSSSAPSS*SSWISGSTATTAR 748
Score = 31.6 bits (70), Expect = 0.39
Identities = 35/169 (20%), Positives = 65/169 (37%)
Frame = +2
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPA 78
TP+P N+ + A ++ TRE + S P S + PS + + +
Sbjct: 380 TPKPGTAKPTPANSTASDAAYIQTPTREPSPESTSTPPASPAKTATLSPSTASSTASRNS 559
Query: 79 ASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWP 138
S P S + R + S+ P+ ST + + S R + +S P + W
Sbjct: 560 HSST*TPTTSPALSRAKSSSSPT------STSSTSATTNSPASFRWMFSSSAPSS*SSWI 721
Query: 139 STMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPS 187
S + + F+ + ++ T++ + R + RQG +PS
Sbjct: 722 SGSTATTARFRRNCLTSTWMSFSSTTTNSPVTSRRISGQ--RRQGISPS 862
>TC9990 similar to UP|Q9FJW0 (Q9FJW0) RuvB DNA helicase-like protein
(AT5g67630/K9I9_20), partial (39%)
Length = 642
Score = 37.0 bits (84), Expect = 0.009
Identities = 37/138 (26%), Positives = 59/138 (41%), Gaps = 12/138 (8%)
Frame = +3
Query: 6 SQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRR 65
SQ ++ + + P PP ++ AT+ A + TS + TPPS+ + S +
Sbjct: 21 SQNPNN*QKPFLRIPLPPRPWRSSSSRRAATSHA*SASAPTPTSAASALTPPSSHATSPK 200
Query: 66 CPSPNLTRTATPAASLQLL--------PKRS-LSAERKRPSTP---PSPPSRRLSTPVND 113
S PA+S + L P S S E RP +P PSP + + +P +
Sbjct: 201 AWSARSPPAKPPASSSR*LRTAKSPVAPSSSPASLEPARPPSPWVWPSPSASKPPSP*SP 380
Query: 114 SAIDGRLSSRKVAASRLP 131
+A RL R++ S P
Sbjct: 381 AASSSRLRCRRLRLSHRP 434
>BP044109
Length = 497
Score = 37.0 bits (84), Expect = 0.009
Identities = 35/135 (25%), Positives = 57/135 (41%), Gaps = 8/135 (5%)
Frame = +2
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPT-PPSTPSGSRRCP------SPNL 71
+P PP KN + A+ S++ T R ++PT PP +P+ S + P P+
Sbjct: 107 SPPPPPPPPNKNKTTPSHASNATSKS---TPRTQTPTTPPPSPTSSPKPPPSTFATKPST 277
Query: 72 TRTATPAASLQ-LLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRL 130
+ TP++S P P++ PSPPS P N S + + A L
Sbjct: 278 SPPETPSSSSHGPAPTLPSPPSSSTPTSTPSPPS-----PPNGSTLPSPPRATPTPAPSL 442
Query: 131 PEGHNLWPSTMRSLS 145
P + +++ S S
Sbjct: 443 PAARRMTSASLCSTS 487
Score = 35.0 bits (79), Expect = 0.035
Identities = 29/96 (30%), Positives = 40/96 (41%), Gaps = 7/96 (7%)
Frame = +2
Query: 309 SLHDLRPHRSSTPHTDRAGLSIAGVRSQSLS----APGSRLPSPSRISVQSTSSSRGVSP 364
SL RP + PHT LS + S S S +P P P++ + +S S
Sbjct: 2 SLQSTRPEQQQPPHTATTSLSHSSSTSLSSSHFFPSPPPPPPPPNKNKTTPSHASNATSK 181
Query: 365 SRSR---PSTPPSRGGVSPSRIRPTSSSIQSNDSVS 397
S R P+TPP SP +P S+ + S S
Sbjct: 182 STPRTQTPTTPPPSPTSSP---KPPPSTFATKPSTS 280
Score = 34.3 bits (77), Expect = 0.060
Identities = 23/77 (29%), Positives = 33/77 (41%)
Frame = +2
Query: 34 TATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAER 93
TAT + S + ++S + P+PP P P PN +T AS + S
Sbjct: 44 TATTSLSHSSSTSLSSSHFFPSPPPPP------PPPNKNKTTPSHAS------NATSKST 187
Query: 94 KRPSTPPSPPSRRLSTP 110
R TP +PP S+P
Sbjct: 188 PRTQTPTTPPPSPTSSP 238
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.125 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,746,956
Number of Sequences: 28460
Number of extensions: 151385
Number of successful extensions: 2755
Number of sequences better than 10.0: 466
Number of HSP's better than 10.0 without gapping: 1988
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2573
length of query: 616
length of database: 4,897,600
effective HSP length: 96
effective length of query: 520
effective length of database: 2,165,440
effective search space: 1126028800
effective search space used: 1126028800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0037a.4


