

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
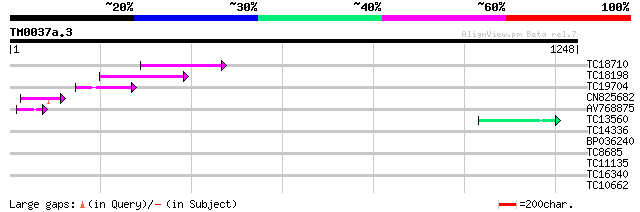
Query= TM0037a.3
(1248 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18710 139 2e-33
TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, p... 132 4e-31
TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA po... 83 3e-16
CN825682 68 1e-11
AV768875 54 2e-07
TC13560 50 2e-06
TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 31 1.1
BP036240 30 1.8
TC8685 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, parti... 30 1.8
TC11135 homologue to UP|SCRK_SOLTU (P37829) Fructokinase , part... 29 4.0
TC16340 weakly similar to UP|Q8S304 (Q8S304) Enoyl-CoA hydratase... 28 8.9
TC10662 homologue to UP|CWP1_YEAST (P28319) Cell wall protein CW... 28 8.9
>TC18710
Length = 843
Score = 139 bits (351), Expect = 2e-33
Identities = 74/190 (38%), Positives = 109/190 (56%), Gaps = 1/190 (0%)
Frame = +2
Query: 289 EIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVL 348
+++ F + + G DG+N LF+Q D + SV I ++R E E+N+TL+ L
Sbjct: 143 KLRLLFFLLETSQPRGMDGMNGLFYQKNGDVVKDSVT*AILAFLERAEIPNEINETLVTL 322
Query: 349 IPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNII 408
IPK+ P ++Q+R I C +YK ++K RLK + DLISP Q FI GRQ DN++
Sbjct: 323 IPKVPHPESINQFRPISGCTFLYKVISKIFVARLKDAMPDLISPMQSGFIQGRQIQDNLL 502
Query: 409 IAQEVFHSM-RYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVS 467
I QE FH++ R + IK+D+ KAYDR+EW FL+ +LL G N+ +I+ VS
Sbjct: 503 IVQEAFHAINRPGALGRNHSIIKLDMNKAYDRVEWKFLESSLLAFGFSTNWVKMIMILVS 682
Query: 468 LSSFQVLFNG 477
S+ NG
Sbjct: 683 GVSYNYKING 712
>TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, partial
(14%)
Length = 592
Score = 132 bits (331), Expect = 4e-31
Identities = 77/195 (39%), Positives = 107/195 (54%), Gaps = 1/195 (0%)
Frame = -3
Query: 199 RVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSP 258
R+ L GD NTRFFH ST+ RR N+I + D HG+ V + A+++F +Y+S
Sbjct: 590 RIK*LK*GDHNTRFFHASTIQRRDFNRILKIKDAHGIWVEGQKKVNEAAVNYFSDIYSSS 411
Query: 259 GTDVLYHVRNAFPTLPRDTLL-EISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQW 317
L P L D L ++ + DEIK AVFSM LKAPG DG+N LF+Q
Sbjct: 410 PLQYLRECLAPIPKLVTDRLNHKLCAQVSTDEIKEAVFSMGDLKAPGMDGINGLFYQQNR 231
Query: 318 DTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKA 377
+ + + V I D E E+N+TL+ LIPKI + Q+R I C+ IYK ++K
Sbjct: 230 EIVKNDVNNAILDFFDHGELPVELNETLVTLIPKIPHAEAIQQFRPISCCSFIYKVISKI 51
Query: 378 LANRLKGVLGDLISP 392
+ RLK + +LISP
Sbjct: 50 IVARLKPDMVNLISP 6
>TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA
polymerase-like protein, partial (3%)
Length = 530
Score = 82.8 bits (203), Expect = 3e-16
Identities = 48/134 (35%), Positives = 65/134 (47%)
Frame = -3
Query: 145 EIGRRKRHLMRRLEGINSRLRMQHIPYLEKLQKRLWKEYQTTLIQEELLWRQKSRVAWLN 204
EI K L NS Q I + K LW+ QEEL W Q+SR+ WL
Sbjct: 384 EIAELKNQLEHLQNYSNSGEDWQEIKQRKMRLKDLWR-------QEELFWSQRSRIKWLK 226
Query: 205 HGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLY 264
GDKNT+FFH ST+ RR+RN+IE + D G V + + DF+ +Y S +
Sbjct: 225 SGDKNTKFFHASTVQRRERNRIERIKDMQGTWVRSQLAINRDVPDFYPDIYKSTNRQNIT 46
Query: 265 HVRNAFPTLPRDTL 278
NA P + ++L
Sbjct: 45 ECLNAVPRVIYESL 4
>CN825682
Length = 626
Score = 67.8 bits (164), Expect = 1e-11
Identities = 40/104 (38%), Positives = 49/104 (46%), Gaps = 5/104 (4%)
Frame = -3
Query: 24 RIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMGKFRDCLDDCYLSDLGFKGPPFTW 83
+ +NT W GDFN L+ +K G + + FRD LD L DL KG FTW
Sbjct: 327 KFKSNTNTFWTCYGDFNEILHHQEKEGARDQDPTRIQVFRDFLDGANLMDLELKGCRFTW 148
Query: 84 -----EGRGVKERLDWALGNDQWLRSFPEASIFHLPQLKSDHKP 122
G KERLD L N W +P A+ LP + SDH P
Sbjct: 147 LSNLRNGLVTKERLDRVLVNWTWRLDYPNATAMALPIISSDHSP 16
>AV768875
Length = 232
Score = 53.9 bits (128), Expect = 2e-07
Identities = 30/70 (42%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Frame = +1
Query: 15 RNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGD-PPNLLAMGKFRDCLDDCYLSD 73
R+ LWQ L I A+ PW+++GD N L GG+ PN +F LDDC L D
Sbjct: 4 RDILWQHLSTIRASISIPWLLIGDMNEILFPSQVRGGEFHPN--RAQRFATVLDDCNLID 177
Query: 74 LGFKGPPFTW 83
LG G FTW
Sbjct: 178LGMVGGKFTW 207
>TC13560
Length = 571
Score = 50.1 bits (118), Expect = 2e-06
Identities = 50/180 (27%), Positives = 71/180 (38%)
Frame = +1
Query: 1032 SHLGFVEVSRNGADSIALSFAPCSIKLTRPSAWSL*LQRWQYCSFRMGEAGCGLGEDQRG 1091
SHLGF E+S GA + ++P+ *L F + + CG+G DQ
Sbjct: 7 SHLGFHEISWYGASGNVFLLESFPVLDSKPAT*IF*L*PNSEHFF*LDKTRCGMG*DQC* 186
Query: 1092 WSFEQ*FNRQLWRCGSQRGGPLDEGFWSSSWHPWCC*RLLRRVDSSTDKIGASHGHGAAA 1151
W +Q L R Q L +GF W+ + C +D D G H G ++
Sbjct: 187 WCGKQLSPG*L*RSAPQCRWELVKGF*MEFWNLFFCQCFPHGIDGCEDGSGGCHEFGLSS 366
Query: 1152 GNC*VRFSRGYQYVKLAFRGSLSPLRAGGLAHPPIAAGPWERYLPAYTEESQLPCRLYS* 1211
NC*+ + R PL++ L + P AA W P Q C L+S*
Sbjct: 367 RNC*IGLLGSC*FSSFN-RPWFPPLQSNRLRYSPPAARAWGSCFP-ICSAGQFNCGLHS* 540
>TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial (85%)
Length = 1917
Score = 31.2 bits (69), Expect = 1.1
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 6/95 (6%)
Frame = -3
Query: 115 QLKSDHKPLLLQLSPIDIDHSQRPNRQVFGEIGRRKRHLM-----RRLEGINSRLR-MQH 168
++ + H+PLLL S I SQ PNR + GE +RH R+ N R R ++H
Sbjct: 1462 KILTQHRPLLLWRS-IGKSRSQSPNRNLLGERSIAERHATDIIGNRKHASSNPRHRNLRH 1286
Query: 169 IPYLEKLQKRLWKEYQTTLIQEELLWRQKSRVAWL 203
P +R+ T +E LLW +++ WL
Sbjct: 1285 TP-----TRRITVH---TAPREHLLWHKRN---WL 1214
>BP036240
Length = 567
Score = 30.4 bits (67), Expect = 1.8
Identities = 14/26 (53%), Positives = 15/26 (56%)
Frame = -2
Query: 1174 SPLRAGGLAHPPIAAGPWERYLPAYT 1199
SP G +P AAGPW LPAYT
Sbjct: 278 SPFWVFGCFYPSYAAGPWLDSLPAYT 201
>TC8685 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, partial (40%)
Length = 1067
Score = 30.4 bits (67), Expect = 1.8
Identities = 22/62 (35%), Positives = 28/62 (44%)
Frame = +1
Query: 18 LWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMGKFRDCLDDCYLSDLGFK 77
L + L +IAA+ P +V GDFN+ G P LLAMGK D + L
Sbjct: 325 LLKGLEKIAASADIPMLVCGDFNS------NPGSAPHALLAMGKVDPSHPDLAIDPLSIL 486
Query: 78 GP 79
P
Sbjct: 487 RP 492
>TC11135 homologue to UP|SCRK_SOLTU (P37829) Fructokinase , partial (33%)
Length = 402
Score = 29.3 bits (64), Expect = 4.0
Identities = 19/50 (38%), Positives = 27/50 (54%)
Frame = +3
Query: 115 QLKSDHKPLLLQLSPIDIDHSQRPNRQVFGEIGRRKRHLMRRLEGINSRL 164
Q + +H L Q SP+ H RP+RQ+ + R + H +RRL SRL
Sbjct: 39 QFRENHG--LQQRSPL---HRHRPHRQLRRDANRLRPHRLRRLPRRGSRL 173
>TC16340 weakly similar to UP|Q8S304 (Q8S304) Enoyl-CoA hydratase, partial
(43%)
Length = 631
Score = 28.1 bits (61), Expect = 8.9
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +3
Query: 1038 EVSRNGADSIALSFAPCSIKLTRPSAWSL*LQRWQYC 1074
++ R G S+ L FAPC+ KL + + + *L +YC
Sbjct: 402 QLPR*GTASLPLPFAPCTGKLPKMKSATF*LCTLRYC 512
>TC10662 homologue to UP|CWP1_YEAST (P28319) Cell wall protein CWP1
precursor, partial (6%)
Length = 937
Score = 28.1 bits (61), Expect = 8.9
Identities = 18/38 (47%), Positives = 20/38 (52%)
Frame = +3
Query: 148 RRKRHLMRRLEGINSRLRMQHIPYLEKLQKRLWKEYQT 185
RR RHL RRL + RLR P LE RL + QT
Sbjct: 246 RRLRHLRRRLIFLLRRLRQIPRPLLEPPPPRLRRHRQT 359
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.343 0.152 0.539
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,273,200
Number of Sequences: 28460
Number of extensions: 401669
Number of successful extensions: 2615
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 2572
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2609
length of query: 1248
length of database: 4,897,600
effective HSP length: 101
effective length of query: 1147
effective length of database: 2,023,140
effective search space: 2320541580
effective search space used: 2320541580
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0037a.3


