

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.6
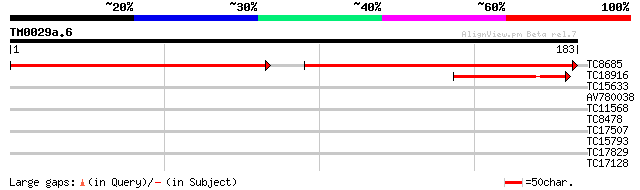
(183 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8685 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, parti... 175 2e-86
TC18916 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, part... 58 8e-10
TC15633 similar to UP|Q8VYU4 (Q8VYU4) At5g11350/F2I11_240, parti... 35 0.008
AV780038 27 1.6
TC11568 similar to UP|Q9AYT8 (Q9AYT8) Mangrin, partial (68%) 27 2.7
TC8478 similar to PIR|B86367|B86367 protein F26F24.16 [imported]... 27 2.7
TC17507 26 4.6
TC15793 similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper protein (AT... 25 7.9
TC17829 similar to UP|Q941D6 (Q941D6) AT4g33470/F17M5_230, parti... 25 7.9
TC17128 homologue to PIR|T06431|T06431 ribosomal protein L27-5 -... 25 7.9
>TC8685 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, partial (40%)
Length = 1067
Score = 175 bits (444), Expect(2) = 2e-86
Identities = 84/84 (100%), Positives = 84/84 (100%)
Frame = +1
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV
Sbjct: 262 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 441
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQ 84
DPSHPDLAIDPLSILRPHSKLVHQ
Sbjct: 442 DPSHPDLAIDPLSILRPHSKLVHQ 513
Score = 159 bits (402), Expect(2) = 2e-86
Identities = 80/88 (90%), Positives = 82/88 (92%)
Frame = +2
Query: 96 RTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRK 155
R IGLG+E RLDSATNEPLFTNVTR+ IGALDYIFYTADSLVVESLLELLDEESLRK
Sbjct: 554 RHIGLGFEHVHCRLDSATNEPLFTNVTRECIGALDYIFYTADSLVVESLLELLDEESLRK 733
Query: 156 DTALPSPEWSSDHIALLAEFRCCKNRSR 183
DTALPSPEWSSDHIALLAEFRCCKNRSR
Sbjct: 734 DTALPSPEWSSDHIALLAEFRCCKNRSR 817
>TC18916 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, partial (4%)
Length = 518
Score = 58.2 bits (139), Expect = 8e-10
Identities = 30/38 (78%), Positives = 31/38 (80%)
Frame = +1
Query: 144 LLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKNR 181
+LELLDEESLRKDT L S EWSSDH A LAEF CC NR
Sbjct: 181 ILELLDEESLRKDTTLHSLEWSSDHDA-LAEFCCCNNR 291
>TC15633 similar to UP|Q8VYU4 (Q8VYU4) At5g11350/F2I11_240, partial (7%)
Length = 660
Score = 35.0 bits (79), Expect = 0.008
Identities = 19/60 (31%), Positives = 31/60 (51%)
Frame = +2
Query: 123 RDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKNRS 182
R F G +DYI + ++ L +L + + + P+ +W SDHIAL++E K S
Sbjct: 5 RCFSGTVDYI-WRSEGLQTTRVLAPIPKHVMEWTPGFPTKKWGSDHIALVSELAFLKEGS 181
>AV780038
Length = 564
Score = 27.3 bits (59), Expect = 1.6
Identities = 12/20 (60%), Positives = 14/20 (70%)
Frame = -3
Query: 78 HSKLVHQLPLVSAYSSFART 97
HS+LVHQLPL S + S T
Sbjct: 379 HSQLVHQLPLPSLFHSHVST 320
>TC11568 similar to UP|Q9AYT8 (Q9AYT8) Mangrin, partial (68%)
Length = 866
Score = 26.6 bits (57), Expect = 2.7
Identities = 14/42 (33%), Positives = 21/42 (49%), Gaps = 2/42 (4%)
Frame = -1
Query: 21 TLLKGLEKIAASADIPMLVCGDFNSN--PGSAPHALLAMGKV 60
T++ +E + IP+L+C FN N P PH L +V
Sbjct: 260 TIVPKVEAVYGFIPIPLLLCFVFNQNTDPCCYPHTFLQASRV 135
>TC8478 similar to PIR|B86367|B86367 protein F26F24.16 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(67%)
Length = 1124
Score = 26.6 bits (57), Expect = 2.7
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = -3
Query: 163 EWSSDHIALLAEFRCCKNRS 182
E+SSDH+ L+ E CK RS
Sbjct: 96 EYSSDHLGLIEEGAQCKTRS 37
>TC17507
Length = 573
Score = 25.8 bits (55), Expect = 4.6
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = +3
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS 32
N H VH+ KD+ L VH LL L +A +
Sbjct: 342 NVHFLVHKAYKDILLTIVHFLLAMLMNVAXN 434
>TC15793 similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper protein
(AT5g58430/mqj2_20), partial (43%)
Length = 969
Score = 25.0 bits (53), Expect = 7.9
Identities = 12/31 (38%), Positives = 18/31 (57%)
Frame = -1
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQL 85
L+ +D + P IDP+SI R + L H+L
Sbjct: 927 LSFSFIDLATPPDGIDPVSIFRNPNTLFHEL 835
>TC17829 similar to UP|Q941D6 (Q941D6) AT4g33470/F17M5_230, partial (30%)
Length = 559
Score = 25.0 bits (53), Expect = 7.9
Identities = 11/27 (40%), Positives = 15/27 (54%)
Frame = +1
Query: 76 RPHSKLVHQLPLVSAYSSFARTIGLGY 102
RPH L+H LP A S + +G G+
Sbjct: 31 RPHLMLLHNLPRSPASGSVSFLVGNGF 111
>TC17128 homologue to PIR|T06431|T06431 ribosomal protein L27-5 - garden pea
{Pisum sativum;}, complete
Length = 576
Score = 25.0 bits (53), Expect = 7.9
Identities = 12/33 (36%), Positives = 19/33 (57%)
Frame = +1
Query: 146 ELLDEESLRKDTALPSPEWSSDHIALLAEFRCC 178
E ++ E +R+ LP+P+ S H +E RCC
Sbjct: 214 EEIEGEGVREARELPAPDAHSLHP*CRSEGRCC 312
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,385,087
Number of Sequences: 28460
Number of extensions: 42139
Number of successful extensions: 196
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 194
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 195
length of query: 183
length of database: 4,897,600
effective HSP length: 85
effective length of query: 98
effective length of database: 2,478,500
effective search space: 242893000
effective search space used: 242893000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0029a.6


