

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.17
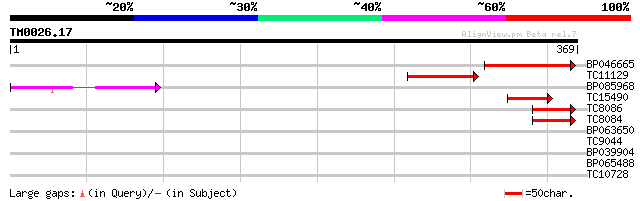
(369 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP046665 75 2e-14
TC11129 weakly similar to UP|PDA6_MEDSA (P38661) Probable protei... 64 3e-11
BP085968 54 3e-08
TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like prote... 44 6e-05
TC8086 similar to UP|DMC1_HUMAN (Q14565) Meiotic recombination p... 41 3e-04
TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombin... 41 3e-04
BP063650 30 0.11
TC9044 similar to GB|AAR21619.1|38490051|AY457043 TATA-binding p... 27 5.4
BP039904 27 7.1
BP065488 26 9.2
TC10728 26 9.2
>BP046665
Length = 524
Score = 75.1 bits (183), Expect = 2e-14
Identities = 44/59 (74%), Positives = 49/59 (82%)
Frame = -2
Query: 310 ILHAQEYQIKKLY*SKVLR*CY*EI*IKQPVYALERG**WLILEQML*RPL**QEPTLE 368
ILHA EYQI K +* K LR*C+*EI IKQ V+A+ER **WLILEQM *+PL** EPTLE
Sbjct: 523 ILHASEYQIIK*H*RKELR*CF*EIYIKQSVFAMERD**WLILEQM**KPL**LEPTLE 347
>TC11129 weakly similar to UP|PDA6_MEDSA (P38661) Probable protein disulfide
isomerase A6 precursor (P5) , partial (7%)
Length = 600
Score = 64.3 bits (155), Expect = 3e-11
Identities = 30/46 (65%), Positives = 38/46 (82%)
Frame = +3
Query: 260 LHWNVLRKLIILCLIYYQIIQQST*AQITLANLTRTLNYNQSGLLQ 305
LHWN+LR+L+ LCLIY+Q I ST*AQI LAN+ +T NY+ +GLLQ
Sbjct: 459 LHWNLLRRLMNLCLIYFQAIPPST*AQIPLANMMKTPNYSHNGLLQ 596
>BP085968
Length = 341
Score = 54.3 bits (129), Expect = 3e-08
Identities = 39/103 (37%), Positives = 48/103 (45%), Gaps = 5/103 (4%)
Frame = +3
Query: 1 FTFCMDMEELAKILSGIHYLNVASSG-----TFSAAIRSMGMIVLNVASSGIASLLLPGG 55
F F MD+E K L GIH L V T+ + +G +
Sbjct: 75 FIFYMDLEVREKHLCGIHGLQV*DHKDSWF*TWHQVVLLLGCCLEE-------------- 212
Query: 56 RTAHSRFCIPLQTDKTATCNIKQDSLRENLLICAKFIIWDEAP 98
RTAHSRF I + + +TCNIKQ S + LL A IIWDEAP
Sbjct: 213 RTAHSRFSIHISINDISTCNIKQGSQKAELLQKASSIIWDEAP 341
>TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like protein, partial
(8%)
Length = 634
Score = 43.5 bits (101), Expect = 6e-05
Identities = 23/29 (79%), Positives = 25/29 (85%)
Frame = +2
Query: 325 KVLR*CY*EI*IKQPVYALERG**WLILE 353
KVL *C+*EI* K PVYA+ER **WLILE
Sbjct: 47 KVL**CF*EI*FKHPVYAMERD**WLILE 133
>TC8086 similar to UP|DMC1_HUMAN (Q14565) Meiotic recombination protein
DMC1/LIM15 homolog, partial (12%)
Length = 628
Score = 41.2 bits (95), Expect = 3e-04
Identities = 22/28 (78%), Positives = 24/28 (85%)
Frame = +2
Query: 341 YALERG**WLILEQML*RPL**QEPTLE 368
YA+ER **WLILE M *+PL** EPTLE
Sbjct: 170 YAMERD**WLILEHMS*KPL**LEPTLE 253
>TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombination
protein PIF1, mitochondrial precursor, partial (4%)
Length = 560
Score = 41.2 bits (95), Expect = 3e-04
Identities = 22/28 (78%), Positives = 24/28 (85%)
Frame = +2
Query: 341 YALERG**WLILEQML*RPL**QEPTLE 368
YA+ER **WLILE M *+PL** EPTLE
Sbjct: 62 YAMERD**WLILEHMS*KPL**LEPTLE 145
>BP063650
Length = 497
Score = 30.4 bits (67), Expect(2) = 0.11
Identities = 15/31 (48%), Positives = 21/31 (67%)
Frame = +1
Query: 141 QILPVIPKGGRQDIVSATVNSSDLWKYCKVL 171
+ L V+ KG QDI+ A VN+S L +C+VL
Sbjct: 58 KFLTVVYKGRMQDIIHAIVNASYL*DHCQVL 150
Score = 20.8 bits (42), Expect(2) = 0.11
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +3
Query: 133 VVLGGDFRQI 142
VVL GDFRQI
Sbjct: 33 VVLQGDFRQI 62
>TC9044 similar to GB|AAR21619.1|38490051|AY457043 TATA-binding protein
associated factor 4 {Arabidopsis thaliana;} , partial
(5%)
Length = 580
Score = 26.9 bits (58), Expect = 5.4
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = +2
Query: 232 LDLIDLHIPIWCRT*RLKSFWKDDAYCVLH 261
L+ + I I C + +LKSF K D CV+H
Sbjct: 362 LNFVVEFIIILCLSTKLKSFLKSDTNCVVH 451
>BP039904
Length = 544
Score = 26.6 bits (57), Expect = 7.1
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = -2
Query: 169 KVLKLTKNMRLCTTGSPEL 187
K+L L +N+RLC TGS L
Sbjct: 384 KILDLLENLRLCVTGSSVL 328
>BP065488
Length = 439
Score = 26.2 bits (56), Expect = 9.2
Identities = 14/17 (82%), Positives = 15/17 (87%)
Frame = -1
Query: 350 LILEQML*RPL**QEPT 366
LILEQM *+PL** EPT
Sbjct: 433 LILEQMS*KPL**LEPT 383
>TC10728
Length = 500
Score = 26.2 bits (56), Expect = 9.2
Identities = 12/32 (37%), Positives = 17/32 (52%)
Frame = -3
Query: 1 FTFCMDMEELAKILSGIHYLNVASSGTFSAAI 32
F +C D+ +K +S +H LNV FS I
Sbjct: 174 FCYCCDVLH*SKSVSHVHNLNVPEVACFSVTI 79
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.342 0.152 0.494
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,067,149
Number of Sequences: 28460
Number of extensions: 83031
Number of successful extensions: 579
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 577
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 579
length of query: 369
length of database: 4,897,600
effective HSP length: 92
effective length of query: 277
effective length of database: 2,279,280
effective search space: 631360560
effective search space used: 631360560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0026.17


