

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
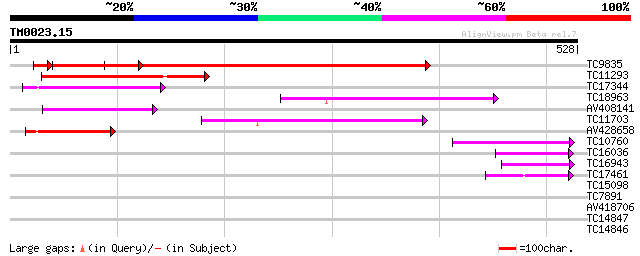
Query= TM0023.15
(528 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9835 similar to AAQ56831 (AAQ56831) At3g03960, partial (67%) 525 e-149
TC11293 homologue to UP|Q8H9B2 (Q8H9B2) T-complex polypeptide 1,... 114 4e-26
TC17344 homologue to GB|AAM26704.1|20857172|AY102137 AT3g11830/F... 99 2e-21
TC18963 homologue to UP|CAE47772 (CAE47772) Cytosolic chaperonin... 92 2e-19
AV408141 89 2e-18
TC11703 homologue to UP|Q9ZSQ6 (Q9ZSQ6) T-complex protein 1 epsi... 64 6e-11
AV428658 60 7e-10
TC10760 similar to UP|Q9ZSQ6 (Q9ZSQ6) T-complex protein 1 epsilo... 60 1e-09
TC16036 homologue to GB|BAB02032.1|9294075|AB020749 cytosolic ch... 47 6e-06
TC16943 similar to UP|O81503 (O81503) F9D12.18 protein, partial ... 47 6e-06
TC17461 similar to UP|Q8L7N0 (Q8L7N0) TCP-1 chaperonin-like prot... 44 7e-05
TC15098 similar to UP|Q8VYV7 (Q8VYV7) AT5g66120/K2A18_20, partia... 29 2.2
TC7891 similar to UP|DCAM_VICFA (Q9M4D8) S-adenosylmethionine de... 28 3.7
AV418706 27 6.3
TC14847 homologue to UP|Q9XG98 (Q9XG98) Phosphoribosyl pyrophosp... 27 6.3
TC14846 homologue to PIR|S71262|S71262 ribose-phosphate diphosph... 27 6.3
>TC9835 similar to AAQ56831 (AAQ56831) At3g03960, partial (67%)
Length = 1126
Score = 525 bits (1351), Expect = e-149
Identities = 277/304 (91%), Positives = 281/304 (92%)
Frame = +1
Query: 89 GKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEK 148
G+ + +G G G + L MGLHPSEIISGYTKAIAKTIEILD LVEK
Sbjct: 214 GRLNRRTLGTGRI*LFHLPGSCCRALRSLSGMGLHPSEIISGYTKAIAKTIEILD*LVEK 393
Query: 149 GSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLVADACIQVCPKNPANFNVDNVRVAK 208
GSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLVADACIQVCPKNPANFNVDNVRVAK
Sbjct: 394 GSESMDVRDKVQVISRMRAAVASKQFGQEDTICSLVADACIQVCPKNPANFNVDNVRVAK 573
Query: 209 LLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTVLIHSAEQLE 268
LLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTVLIHSAEQLE
Sbjct: 574 LLGGGLHNSTVVRGMVLKSDAVGSIKRIEKAKVAVFVSGVDTSATDTKGTVLIHSAEQLE 753
Query: 269 NYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFELRRFCR 328
NYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFELRRFCR
Sbjct: 754 NYSKTEEAKVEELIKAVADSGAKVIVSGGAVGEMALHFCERYKLMVLKISSKFELRRFCR 933
Query: 329 TTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGSTDSILD 388
TTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGSTDSILD
Sbjct: 934 TTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKNEEGGNSVSTVVLRGSTDSILD 1113
Query: 389 DLER 392
DLER
Sbjct: 1114DLER 1125
Score = 156 bits (395), Expect(2) = 3e-44
Identities = 80/84 (95%), Positives = 81/84 (96%)
Frame = +3
Query: 41 ITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAGKAQQEEIGDGA 100
ITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAGKAQQE+IGDGA
Sbjct: 69 ITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAGKAQQEDIGDGA 248
Query: 101 NLTISFAGELLQGAEELIRMGLHP 124
NLTISFAGELLQGAEELIR G P
Sbjct: 249 NLTISFAGELLQGAEELIRDGFAP 320
Score = 39.3 bits (90), Expect(2) = 3e-44
Identities = 18/18 (100%), Positives = 18/18 (100%)
Frame = +2
Query: 23 GLDEAVIKNIDACKQLST 40
GLDEAVIKNIDACKQLST
Sbjct: 14 GLDEAVIKNIDACKQLST 67
>TC11293 homologue to UP|Q8H9B2 (Q8H9B2) T-complex polypeptide 1, partial
(32%)
Length = 591
Score = 114 bits (285), Expect = 4e-26
Identities = 59/158 (37%), Positives = 99/158 (62%), Gaps = 1/158 (0%)
Frame = +3
Query: 30 KNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAG 89
+N+ AC+ ++ I +TSLGP G++KM+++ + + +TND ATI+ LEV+HPAAK+LV
Sbjct: 126 QNVVACQAVANIVKTSLGPVGLDKMLVDDIGDVTITNDGATILRMLEVEHPAAKVLVELA 305
Query: 90 KAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAKTIEILDELVEKG 149
+ Q E+GDG + A ELL+ A +L+R +HP+ IISGY A+ + + +D EK
Sbjct: 306 ELQDREVGDGTTSVVIVAAELLKRANDLVRNKIHPTSIISGYRLAMREACKYID---EKL 476
Query: 150 SESMDVRDKVQVISRMRAAVASKQF-GQEDTICSLVAD 186
+ +D K +++ + +++SK G D ++V D
Sbjct: 477 AVKVDKLGKDSLVNCAKTSMSSKLIAGDSDFFANMVVD 590
>TC17344 homologue to GB|AAM26704.1|20857172|AY102137 AT3g11830/F26K24_12
{Arabidopsis thaliana;}, partial (28%)
Length = 567
Score = 98.6 bits (244), Expect = 2e-21
Identities = 50/133 (37%), Positives = 79/133 (58%)
Frame = +1
Query: 13 MLKEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIV 72
+LKEG G + ++ NI+AC ++ + RT+LGP GM+K++ + + ++ND ATI+
Sbjct: 124 LLKEGTDTSQGKAQ-LVSNINACTAVADVVRTTLGPRGMDKLIHDDKGSVTISNDGATIM 300
Query: 73 NELEVQHPAAKILVLAGKAQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYT 132
L++ HPAAKILV K+Q E+GDG + A E L+ A+ I G+HP +I Y
Sbjct: 301 KLLDIVHPAAKILVDIAKSQDSEVGDGTTTVVLLAAEFLREAKPFIEDGVHPQNLIRSYR 480
Query: 133 KAIAKTIEILDEL 145
A I+ + +L
Sbjct: 481 TASTLAIQKVKDL 519
>TC18963 homologue to UP|CAE47772 (CAE47772) Cytosolic chaperonin
delta-subunit, partial (41%)
Length = 652
Score = 92.4 bits (228), Expect = 2e-19
Identities = 51/208 (24%), Positives = 106/208 (50%), Gaps = 5/208 (2%)
Frame = +1
Query: 253 TDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVI-----VSGGAVGEMALHFC 307
TD + ++++ Q++ K E + + +IK + +G V+ + AV +++LH+
Sbjct: 28 TDIEQSIVVSDYSQMDRILKEERSYILGMIKKIKATGCNVLLIQKSILRDAVTDLSLHYL 207
Query: 308 ERYKLMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEEVGGVTVTIVKN 367
+ K++V+K + E+ +T + + + + LGY D V +G + +
Sbjct: 208 AKAKILVIKDVERDEIEFITKTLNCLPIANIEHFRAEKLGYADLVEEVSLGDGKIVKISG 387
Query: 368 EEGGNSVSTVVLRGSTDSILDDLERAVDDGVNTYKAMCKDSRTVPGAAATEIELAKRVKD 427
+ +TV++RGS +LD+ ER++ D + + + + G A EIEL++++
Sbjct: 388 IKDMGRTTTVLVRGSNQLVLDEAERSLHDALCVVRCLVAKRFLIAGGGAPEIELSRQLGA 567
Query: 428 FSFKETGLDQYAIAKFAESFEMIPRTLA 455
++ G++ Y + FAE+ E+IP TLA
Sbjct: 568 WAKVLHGMEGYCVRAFAEALEVIPYTLA 651
>AV408141
Length = 427
Score = 89.0 bits (219), Expect = 2e-18
Identities = 45/107 (42%), Positives = 64/107 (59%)
Frame = +1
Query: 31 NIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNELEVQHPAAKILVLAGK 90
NI A + ++ RTSLGP GM+KM+ D++ +TND ATI+N+++V PAAK+LV K
Sbjct: 106 NIVAARSVANAVRTSLGPKGMDKMISTSSDEVIITNDGATILNKMQVLQPAAKMLVELSK 285
Query: 91 AQQEEIGDGANLTISFAGELLQGAEELIRMGLHPSEIISGYTKAIAK 137
+Q GDG + AG LL+ L+ G+HP+ I KA K
Sbjct: 286 SQDSAAGDGTTTVVVIAGALLEQCLLLLSRGIHPTVISDSLYKASIK 426
>TC11703 homologue to UP|Q9ZSQ6 (Q9ZSQ6) T-complex protein 1 epsilon subunit
(Fragment), partial (53%)
Length = 665
Score = 63.9 bits (154), Expect = 6e-11
Identities = 47/214 (21%), Positives = 93/214 (42%), Gaps = 3/214 (1%)
Frame = +1
Query: 179 TICSLVADACIQVCPKNPANFNVDNVRVAKLLGGGLHNSTVVRGMVLKSDA--VGSIKRI 236
++ + +A + V + N+D ++V +GG L ++ ++ G+V+ D K+I
Sbjct: 22 SLAEIAVNAVVAVADLERKDVNLDLIKVEGKVGGKLEDTELIYGIVVDKDMSHPQMPKQI 201
Query: 237 EKAKVAVFVSGVDTSATDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSG 296
E AK+A+ + TK V I + E+ + K E +++++ D+GA +++
Sbjct: 202 EDAKIAILTCPFEPPKPKTKHKVDIDTVEKFQTLRKQEVKYFDDMVQQCKDAGATLVICQ 381
Query: 297 GAVGEMALHFCERYKLMVLKISSKFELRRFCRTTGAVAVLKLSQPNPDDLGYVDSVSVEE 356
+ A H L ++ EL TG V + + P+ LG V +
Sbjct: 382 WGFDDEANHLLMHRNLPAVRWVGGVELELIAIATGGRIVPRFQELTPEKLGKAGLVREKA 561
Query: 357 VGGVTVTIVKNEEGGNS-VSTVVLRGSTDSILDD 389
G ++ E NS T+ +RG I+++
Sbjct: 562 FGTTKDRMLYIEHCANSRAVTIFIRGGNKMIIEE 663
>AV428658
Length = 392
Score = 60.5 bits (145), Expect = 7e-10
Identities = 35/84 (41%), Positives = 52/84 (61%)
Frame = +3
Query: 15 KEGHKHLSGLDEAVIKNIDACKQLSTITRTSLGPNGMNKMVINHLDKLFVTNDAATIVNE 74
+E L GLD A NI A K ++ I RTSLGP GM+KM+ + + +TND ATI+ +
Sbjct: 144 QEQKSRLRGLD-AQKANIFAGKAVARILRTSLGPKGMDKMLQSPDGDVTITNDGATILEQ 320
Query: 75 LEVQHPAAKILVLAGKAQQEEIGD 98
++V + AK++V ++ EIGD
Sbjct: 321 MDVDNQIAKLMVELSRSHDYEIGD 392
>TC10760 similar to UP|Q9ZSQ6 (Q9ZSQ6) T-complex protein 1 epsilon subunit
(Fragment), partial (28%)
Length = 539
Score = 59.7 bits (143), Expect = 1e-09
Identities = 34/115 (29%), Positives = 62/115 (53%), Gaps = 1/115 (0%)
Frame = +1
Query: 413 GAAATEIELAKRVKDFSFKETGLDQYAIAKFAESFEMIPRTLAENAGLNAMEIISSLYAE 472
G + EI + V+ + + G++QYAI FA++ E +P LAEN+GL +E +S++ ++
Sbjct: 1 GGGSAEISCSVAVEAAADRFPGVEQYAIRAFADALEYVPMALAENSGLQPIETLSAVKSQ 180
Query: 473 H-ASGNTKVGIDLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCTVLRVDQVIN 526
N GID + D+ +V++ + K + A V +L++D VI+
Sbjct: 181 QIKDNNPHYGIDCNDVGTNDMREQNVFETLIGKQQQILLATQVVKMILKIDDVIS 345
>TC16036 homologue to GB|BAB02032.1|9294075|AB020749 cytosolic chaperonin,
delta-subunit {Arabidopsis thaliana;} , partial (14%)
Length = 575
Score = 47.4 bits (111), Expect = 6e-06
Identities = 23/73 (31%), Positives = 40/73 (54%)
Frame = +2
Query: 453 TLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDLHVTKFFALKYAA 512
TLAENAGLN + I++ L HA G GI++ +G ++ +V + A+ A
Sbjct: 2 TLAENAGLNPIAIVTELRNRHAQGEINTGINVRKGQITNILEENVVQPLLVSTSAITLAT 181
Query: 513 DAVCTVLRVDQVI 525
+ V +L++D ++
Sbjct: 182 ECVRMILKIDDIV 220
>TC16943 similar to UP|O81503 (O81503) F9D12.18 protein, partial (18%)
Length = 624
Score = 47.4 bits (111), Expect = 6e-06
Identities = 22/69 (31%), Positives = 38/69 (54%), Gaps = 1/69 (1%)
Frame = +2
Query: 459 GLNAMEIISSLYAEHASG-NTKVGIDLEEGVCKDVTTTHVWDLHVTKFFALKYAADAVCT 517
G+N + +++L HA+G N +GID G D+ +WD + K K A +A C
Sbjct: 5 GVNVIXTMTALQGTHANGENAWIGIDGNTGALADMKERKIWDAYNVKAPTFKTALEAACM 184
Query: 518 VLRVDQVIN 526
+LR+D +++
Sbjct: 185 LLRIDDIVS 211
>TC17461 similar to UP|Q8L7N0 (Q8L7N0) TCP-1 chaperonin-like protein
(At5g16070), partial (17%)
Length = 606
Score = 43.9 bits (102), Expect = 7e-05
Identities = 27/82 (32%), Positives = 43/82 (51%)
Frame = +3
Query: 444 AESFEMIPRTLAENAGLNAMEIISSLYAEHASGNTKVGIDLEEGVCKDVTTTHVWDLHVT 503
A++ ++P+TLAEN+GL+ ++I SL EH GN VG+ G D ++D +
Sbjct: 3 ADALLVVPKTLAENSGLDTQDVIISLTGEHDRGNV-VGLCQNTGEPIDPQMEGIFDHYAV 179
Query: 504 KFFALKYAADAVCTVLRVDQVI 525
K + +L VD+VI
Sbjct: 180 KRQIINSGPVIASQLLLVDEVI 245
>TC15098 similar to UP|Q8VYV7 (Q8VYV7) AT5g66120/K2A18_20, partial (66%)
Length = 1272
Score = 28.9 bits (63), Expect = 2.2
Identities = 16/35 (45%), Positives = 21/35 (59%)
Frame = -1
Query: 292 VIVSGGAVGEMALHFCERYKLMVLKISSKFELRRF 326
VIVSGGA+G +A C + L L I S + RR+
Sbjct: 720 VIVSGGAIGNLA---CIKMSLTRLTIESSIQPRRY 625
>TC7891 similar to UP|DCAM_VICFA (Q9M4D8) S-adenosylmethionine
decarboxylase proenzyme (AdoMetDC) (SamDC) [Contains:
S-adenosylmethionine decarboxylase alpha chain;
S-adenosylmethionine decarboxylase beta chain] ,
complete
Length = 1889
Score = 28.1 bits (61), Expect = 3.7
Identities = 18/66 (27%), Positives = 30/66 (45%)
Frame = +1
Query: 240 KVAVFVSGVDTSATDTKGTVLIHSAEQLENYSKTEEAKVEELIKAVADSGAKVIVSGGAV 299
K+A FVS S T+G+ + A+ + S +EE + + SG+ + GG+
Sbjct: 832 KLAEFVSLNVKSVRYTRGSFIFPGAQSFPHRSFSEEVAILDGYFGKLGSGSSAYIMGGSD 1011
Query: 300 GEMALH 305
E H
Sbjct: 1012SEQKWH 1029
>AV418706
Length = 374
Score = 27.3 bits (59), Expect = 6.3
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 4/50 (8%)
Frame = -1
Query: 264 AEQLENYSKTEEAKVEEL----IKAVADSGAKVIVSGGAVGEMALHFCER 309
AE+ + + E+ EE + A A G GGAVG + L FC R
Sbjct: 218 AEEDDEDEEDEDEPAEECAAATVAAAAARGCSTGTVGGAVGAVELGFCRR 69
>TC14847 homologue to UP|Q9XG98 (Q9XG98) Phosphoribosyl pyrophosphate
synthase , partial (75%)
Length = 756
Score = 27.3 bits (59), Expect = 6.3
Identities = 15/35 (42%), Positives = 19/35 (53%), Gaps = 1/35 (2%)
Frame = +1
Query: 238 KAKVAVFVSG-VDTSATDTKGTVLIHSAEQLENYS 271
K KVAV V +DT+ T KG L+H E Y+
Sbjct: 520 KGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYA 624
>TC14846 homologue to PIR|S71262|S71262 ribose-phosphate diphosphokinase
isoform II [validated] - Arabidopsis
thaliana (fragment) {Arabidopsis thaliana;} , partial
(55%)
Length = 689
Score = 27.3 bits (59), Expect = 6.3
Identities = 15/35 (42%), Positives = 19/35 (53%), Gaps = 1/35 (2%)
Frame = +1
Query: 238 KAKVAVFVSG-VDTSATDTKGTVLIHSAEQLENYS 271
K KVAV V +DT+ T KG L+H E Y+
Sbjct: 211 KGKVAVMVDDMIDTAGTIAKGAALLHEEGAREVYA 315
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,467,967
Number of Sequences: 28460
Number of extensions: 66630
Number of successful extensions: 336
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 336
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 336
length of query: 528
length of database: 4,897,600
effective HSP length: 94
effective length of query: 434
effective length of database: 2,222,360
effective search space: 964504240
effective search space used: 964504240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0023.15


