

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
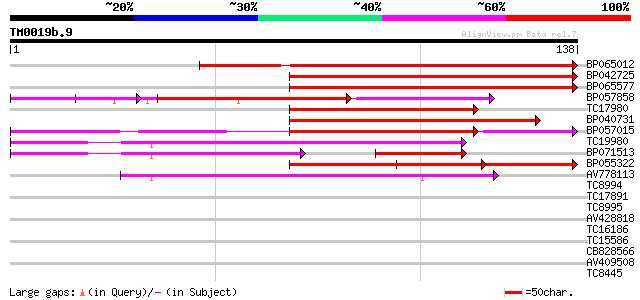
Query= TM0019b.9
(138 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP065012 103 1e-23
BP042725 102 2e-23
BP065577 96 2e-21
BP057858 76 1e-18
TC17980 79 3e-16
BP040731 74 9e-15
BP057015 74 1e-14
TC19980 similar to UP|Q9FNP7 (Q9FNP7) Emb|CAB71880.1, partial (8%) 71 8e-14
BP071513 60 2e-12
BP055322 59 4e-10
AV778113 58 7e-10
TC8994 homologue to UP|RS0_NEUCR (Q01291) 40S ribosomal protein ... 37 0.002
TC17891 33 0.018
TC8995 32 0.051
AV428818 30 0.11
TC16186 weakly similar to UP|Q9LTB9 (Q9LTB9) Genomic DNA, chromo... 30 0.11
TC15586 weakly similar to UP|Q8LEJ2 (Q8LEJ2) Nematode resistance... 30 0.11
CB828566 30 0.15
AV409508 29 0.26
TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial ... 29 0.33
>BP065012
Length = 476
Score = 103 bits (257), Expect = 1e-23
Identities = 62/92 (67%), Positives = 65/92 (70%)
Frame = +3
Query: 47 QNRHTPYDLHIGEPQESNAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVF 106
Q RHTP I S A TRN KYKFLKSLCK KPI VS LT SSPTP SILRV
Sbjct: 57 QVRHTPKGK*IYG*TWSKAS--TRNPKYKFLKSLCKLKPIRVSFLTQDSSPTPDSILRVI 230
Query: 107 HNSSSPHLQLAQRSIFIEDSPSPNSVKRPNNK 138
H+SSS L+L +RS IEDSPSPNSVKRPNNK
Sbjct: 231 HSSSSSPLRLVRRSGSIEDSPSPNSVKRPNNK 326
>BP042725
Length = 539
Score = 102 bits (255), Expect = 2e-23
Identities = 53/70 (75%), Positives = 58/70 (82%)
Frame = +2
Query: 69 TRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQLAQRSIFIEDSPS 128
TRN KYKFLKSLC+ KPI V SLT GS+PTP SIL+VFH+SSS L L +RS IEDSPS
Sbjct: 104 TRNPKYKFLKSLCRLKPIMVRSLTQGSNPTPDSILQVFHSSSSSPL*LVRRSSSIEDSPS 283
Query: 129 PNSVKRPNNK 138
PNSVKRPNNK
Sbjct: 284 PNSVKRPNNK 313
Score = 26.6 bits (57), Expect = 1.7
Identities = 12/22 (54%), Positives = 16/22 (72%)
Frame = +3
Query: 1 MSCILPTVNKFTSSLETRQVPE 22
M C+LPTVN+ + TR+VPE
Sbjct: 45 M*CLLPTVNRIYITP*TRRVPE 110
>BP065577
Length = 301
Score = 96.3 bits (238), Expect = 2e-21
Identities = 51/70 (72%), Positives = 57/70 (80%)
Frame = +2
Query: 69 TRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQLAQRSIFIEDSPS 128
TRN KYKFLKSLC+ KPITVSSLT G + T SIL+VFH+SSS L+L +RS IEDSPS
Sbjct: 68 TRNPKYKFLKSLCQLKPITVSSLTQGFNSTXDSILQVFHSSSSSPLRLVRRSSSIEDSPS 247
Query: 129 PNSVKRPNNK 138
NSVKRPNNK
Sbjct: 248 XNSVKRPNNK 277
Score = 32.0 bits (71), Expect = 0.039
Identities = 14/25 (56%), Positives = 18/25 (72%)
Frame = +1
Query: 1 MSCILPTVNKFTSSLETRQVPEGKL 25
MSCILPT FTSSLE + P+ ++
Sbjct: 10 MSCILPTTKGFTSSLEQGEYPKPQI 84
>BP057858
Length = 382
Score = 76.3 bits (186), Expect(2) = 1e-18
Identities = 35/47 (74%), Positives = 39/47 (82%)
Frame = +2
Query: 37 QQEIHYAKAPQNRHTPYDLHIGEPQESNAPEPTRNLKYKFLKSLCKP 83
QQ +YA P R+TPYDLHIGEPQESNA EPTRNLKYKF+KSL +P
Sbjct: 134 QQRTYYAMTP*IRNTPYDLHIGEPQESNASEPTRNLKYKFIKSLSQP 274
Score = 56.2 bits (134), Expect = 2e-09
Identities = 46/114 (40%), Positives = 55/114 (47%), Gaps = 12/114 (10%)
Frame = +3
Query: 17 TRQVPEGK--LKTHTKPN------KNQIQQEIHYAKAPQNRHTPYD----LHIGEPQESN 64
TR+VPE K + TKPN K I Q H ++ T + +H P+ SN
Sbjct: 66 TRRVPESKPEITQPTKPNQ*KQLNKEPIMQ*HHKSEILPTISTSGNRRKAMHRNLPETSN 245
Query: 65 APEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQLAQ 118
L K PITVSSLT S P P SIL VFHNSSSPHL+L +
Sbjct: 246 INS*NHYLNLK---------PITVSSLTQXSIPRPDSIL*VFHNSSSPHLELVR 380
Score = 31.2 bits (69), Expect(2) = 1e-18
Identities = 16/32 (50%), Positives = 19/32 (59%)
Frame = +1
Query: 1 MSCILPTVNKFTSSLETRQVPEGKLKTHTKPN 32
MSCILPT FTSSLE + P+ K+ N
Sbjct: 19 MSCILPTTMGFTSSLEQGEYPKVNPKSPNLQN 114
>TC17980
Length = 425
Score = 79.0 bits (193), Expect = 3e-16
Identities = 39/46 (84%), Positives = 41/46 (88%)
Frame = -1
Query: 69 TRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHL 114
TRN KYKFLKSLC+ KPITVSSLT GSSPTP SILRV H+SSSPHL
Sbjct: 308 TRNPKYKFLKSLCELKPITVSSLTQGSSPTPDSILRVIHSSSSPHL 171
Score = 34.3 bits (77), Expect = 0.008
Identities = 14/22 (63%), Positives = 18/22 (81%)
Frame = -2
Query: 1 MSCILPTVNKFTSSLETRQVPE 22
MSCILPT +FTS+LE ++PE
Sbjct: 367 MSCILPTTKRFTSNLEQDELPE 302
>BP040731
Length = 492
Score = 73.9 bits (180), Expect = 9e-15
Identities = 42/61 (68%), Positives = 45/61 (72%)
Frame = +2
Query: 69 TRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQLAQRSIFIEDSPS 128
TRN K KFL S CK KPITVSSLT GSSPT SILRV H SSS L+L +RS IE+S S
Sbjct: 77 TRNPK*KFLNSWCKLKPITVSSLTQGSSPTHDSILRVIHISSSSSLRLVRRSSSIENSSS 256
Query: 129 P 129
P
Sbjct: 257 P 259
Score = 32.7 bits (73), Expect = 0.023
Identities = 20/53 (37%), Positives = 28/53 (52%)
Frame = +1
Query: 1 MSCILPTVNKFTSSLETRQVPEGKLKTHTKPNKNQIQQEIHYAKAPQNRHTPY 53
MSCILPT FTSSLE + P+ +++ +K +Q + K R PY
Sbjct: 19 MSCILPTTKGFTSSLEQGEYPKPQIEI----SKLMVQA*TNNGKLSNPRL*PY 165
>BP057015
Length = 450
Score = 73.6 bits (179), Expect = 1e-14
Identities = 38/46 (82%), Positives = 38/46 (82%)
Frame = +1
Query: 69 TRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHL 114
TRN KYKFLKSLCK KPITVSSLT G SPTP S LRVFH SSSP L
Sbjct: 103 TRNPKYKFLKSLCKLKPITVSSLTQGFSPTPDSNLRVFHRSSSPPL 240
Score = 49.7 bits (117), Expect = 2e-07
Identities = 48/138 (34%), Positives = 58/138 (41%)
Frame = +3
Query: 1 MSCILPTVNKFTSSLETRQVPEGKLKTHTKPNKNQIQQEIHYAKAPQNRHTPYDLHIGEP 60
MSCIL TVNKFTS+LE + P+ ++ +K +Q + K R PY
Sbjct: 45 MSCILSTVNKFTSNLEQGEYPKPQI*I----SKIIMQT*TNNGKLSNPRL*PY------- 191
Query: 61 QESNAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQLAQRS 120
+ L S P + SSSP LAQR
Sbjct: 192 ----------------------------TRL*SSSLPQVF-------KSSSP---LAQRL 257
Query: 121 IFIEDSPSPNSVKRPNNK 138
FIEDSP PNSVKRPNNK
Sbjct: 258 SFIEDSPLPNSVKRPNNK 311
>TC19980 similar to UP|Q9FNP7 (Q9FNP7) Emb|CAB71880.1, partial (8%)
Length = 540
Score = 70.9 bits (172), Expect = 8e-14
Identities = 49/112 (43%), Positives = 59/112 (51%), Gaps = 1/112 (0%)
Frame = -2
Query: 1 MSCILPTVNKFTSSLETRQVPEGKLKTHTKPNK-NQIQQEIHYAKAPQNRHTPYDLHIGE 59
M C+L TVN +L Q H +K N+I Q HYA P R+TPYD H G
Sbjct: 497 M*CMLSTVNNLHLALTKAQ--------HKGQDK*NKI*QRTHYAMTP*IRYTPYDFHTG* 342
Query: 60 PQESNAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSS 111
P ESNAPEPT+NLK + + L GSS TP SI RVF+ SS+
Sbjct: 341 P*ESNAPEPTQNLKCES*NYPTSLNQ*R*AILPKGSSLTPDSIFRVFYRSST 186
>BP071513
Length = 410
Score = 59.7 bits (143), Expect(2) = 2e-12
Identities = 36/73 (49%), Positives = 41/73 (55%), Gaps = 1/73 (1%)
Frame = +3
Query: 1 MSCILPTVNKFTSSLETRQVPEGKLKTHTKPNK-NQIQQEIHYAKAPQNRHTPYDLHIGE 59
M CILPTVN +L Q H +K N+I Q HYA P R+TPYD H G
Sbjct: 60 M*CILPTVNNLHLALTKAQ--------HKGQDK*NKI*QRTHYAMTP*IRYTPYDFHTG* 215
Query: 60 PQESNAPEPTRNL 72
P ESNAPEPT+ L
Sbjct: 216 P*ESNAPEPTQRL 254
Score = 26.6 bits (57), Expect(2) = 2e-12
Identities = 13/22 (59%), Positives = 17/22 (77%)
Frame = +1
Query: 90 SLTHGSSPTPYSILRVFHNSSS 111
+L GSS TP SILRVF++ S+
Sbjct: 238 NLPKGSSLTPDSILRVFYSPST 303
>BP055322
Length = 512
Score = 58.5 bits (140), Expect = 4e-10
Identities = 31/48 (64%), Positives = 34/48 (70%)
Frame = +3
Query: 69 TRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQL 116
T+N KYKFLKSLC+ KP VSSLT GSS P S +V H SSPHL L
Sbjct: 111 TQNPKYKFLKSLCRLKPNMVSSLTQGSSLKPDSTRQVLHCPSSPHLYL 254
Score = 42.0 bits (97), Expect = 4e-05
Identities = 27/44 (61%), Positives = 28/44 (63%)
Frame = +2
Query: 95 SSPTPYSILRVFHNSSSPHLQLAQRSIFIEDSPSPNSVKRPNNK 138
SS TP S F +SS LAQR EDSPSPNSVKRPNNK
Sbjct: 212 SSSTPLS----FKSSSL----LAQRLSSFEDSPSPNSVKRPNNK 319
Score = 26.2 bits (56), Expect = 2.2
Identities = 11/22 (50%), Positives = 17/22 (77%)
Frame = +1
Query: 1 MSCILPTVNKFTSSLETRQVPE 22
M C+LPTVN+ ++ TR+VP+
Sbjct: 52 M*CLLPTVNRIYNTP*TRRVPK 117
>AV778113
Length = 553
Score = 57.8 bits (138), Expect = 7e-10
Identities = 39/97 (40%), Positives = 50/97 (51%), Gaps = 5/97 (5%)
Frame = +1
Query: 28 HTKPNK-NQIQQEIHYAKAPQNRHTPYDLHIGEPQESNAPEPTRNLKYKFLKSLCKPKPI 86
H + +K N+IQQ HYA P R+T YD H P ES APEPT+NLKYK
Sbjct: 73 HKRQDK*NKIQQRTHYAMTP*IRYTQYDFHTV*P*ESQAPEPTQNLKYKS*NDPTSLNQ* 252
Query: 87 TVSSLTHGSSPTP----YSILRVFHNSSSPHLQLAQR 119
+ L GSS TP S + + H +S H+ +R
Sbjct: 253 R*AILAKGSSLTPTLFFESSIILPHGVASAHISTFRR 363
>TC8994 homologue to UP|RS0_NEUCR (Q01291) 40S ribosomal protein S0
(Ribosome-associated protein 1), partial (6%)
Length = 1009
Score = 36.6 bits (83), Expect = 0.002
Identities = 25/65 (38%), Positives = 38/65 (58%)
Frame = -2
Query: 34 NQIQQEIHYAKAPQNRHTPYDLHIGEPQESNAPEPTRNLKYKFLKSLCKPKPITVSSLTH 93
N++Q I P ++T Y+ G+ Q+SNA EPT++ + K+ K +P+TVS T
Sbjct: 663 NKVQNVI----TP*VQYTSYNPLTGDSQKSNASEPTQD-QSKYYNLPYKLQPMTVSFSTQ 499
Query: 94 GSSPT 98
GSS T
Sbjct: 498 GSSLT 484
>TC17891
Length = 487
Score = 33.1 bits (74), Expect = 0.018
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = -3
Query: 124 EDSPSPNSVKRPNNK 138
E SPSPNSVKRPNNK
Sbjct: 218 EGSPSPNSVKRPNNK 174
>TC8995
Length = 560
Score = 31.6 bits (70), Expect = 0.051
Identities = 25/72 (34%), Positives = 38/72 (52%)
Frame = -2
Query: 39 EIHYAKAPQNRHTPYDLHIGEPQESNAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPT 98
++ A P ++T Y+ G+ Q+SNA EPT++ + K + +P+TVS T SS T
Sbjct: 355 KVQNAITP*VQYTSYNPLTGDSQKSNASEPTQD-QSK*YNHSYELQPMTVSFSTQDSSLT 179
Query: 99 PYSILRVFHNSS 110
FH SS
Sbjct: 178 SN---YYFHKSS 152
>AV428818
Length = 417
Score = 30.4 bits (67), Expect = 0.11
Identities = 38/129 (29%), Positives = 51/129 (39%), Gaps = 1/129 (0%)
Frame = +3
Query: 4 ILP-TVNKFTSSLETRQVPEGKLKTHTKPNKNQIQQEIHYAKAPQNRHTPYDLHIGEPQE 62
+LP T+N F S + R V L T P+ Q Q RH P P
Sbjct: 9 LLPKTLNLFPSPPQWRPV-SATLSAATPPSSPQTHQ----------RH*PQRRSQEPPST 155
Query: 63 SNAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQLAQRSIF 122
S+ P+PTRN S P++ + T +SP+P S SP+ + A S
Sbjct: 156 SSNPKPTRN-----ASSTSAAPPLSPRTSTSTASPSP---------SPSPNSKTATTS-- 287
Query: 123 IEDSPSPNS 131
PS NS
Sbjct: 288 ---PPSTNS 305
>TC16186 weakly similar to UP|Q9LTB9 (Q9LTB9) Genomic DNA, chromosome 5, TAC
clone:K14A3 (AT5g47090/K14A3_4), partial (54%)
Length = 549
Score = 30.4 bits (67), Expect = 0.11
Identities = 29/96 (30%), Positives = 41/96 (42%), Gaps = 4/96 (4%)
Frame = -3
Query: 46 PQNRHTPYDLHIGEPQE----SNAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPTPYS 101
P RHTP L + P S AP P+ + +S P P GSSP+P+S
Sbjct: 475 PGPRHTPPTLILKPPPHTHDASKAPSPSSPPR----RSTAHPSPTYPGKHAAGSSPSPWS 308
Query: 102 ILRVFHNSSSPHLQLAQRSIFIEDSPSPNSVKRPNN 137
SS+P + RS + SP + RP++
Sbjct: 307 -------SSAPPPSDSSRSAAASRANSPRN--RPSS 227
>TC15586 weakly similar to UP|Q8LEJ2 (Q8LEJ2) Nematode resistance
protein-like protein, partial (32%)
Length = 773
Score = 30.4 bits (67), Expect = 0.11
Identities = 28/97 (28%), Positives = 38/97 (38%), Gaps = 2/97 (2%)
Frame = +1
Query: 6 PTVNKFTSSLETRQVPEGKL--KTHTKPNKNQIQQEIHYAKAPQNRHTPYDLHIGEPQES 63
PT + TS TR E L + T Q I + Q R PY L +GEP +
Sbjct: 502 PTADLTTSETSTRSYSETSLLPRLATWYRSKDAAQRILLSVECQMRRCPYTLGLGEPNLA 681
Query: 64 NAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPTPY 100
P +L Y ++C+P + H TPY
Sbjct: 682 AKP----SLMY---DAVCRPNEV------HELKTTPY 753
>CB828566
Length = 543
Score = 30.0 bits (66), Expect = 0.15
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Frame = +2
Query: 83 PKPITVSSLTHGSSPTPYSILRVFHNSSSPHLQLAQRSIF-IEDSPSPNS 131
P P T+++ H P+P+SI H S SP +L S F ++D P S
Sbjct: 8 PSPFTLAARHHAVMPSPFSI--AVHASCSPSRRLRSPSPFSLQDHPQAYS 151
>AV409508
Length = 426
Score = 29.3 bits (64), Expect = 0.26
Identities = 27/100 (27%), Positives = 39/100 (39%), Gaps = 6/100 (6%)
Frame = +2
Query: 41 HYAKAPQN--RHTPYDLHIGEPQESNAPEPTRNLKYKFLKSLCKPKPITVSSLTHGSSPT 98
H PQ+ RHT + L P S P P L + +LK + TH + T
Sbjct: 107 HTLSIPQSNSRHTDFPLKQSNPPSSPPPPPPPTLLFHYLKKI---------QTTHLFNST 259
Query: 99 PYSILRVFHNSSSPHL----QLAQRSIFIEDSPSPNSVKR 134
P S V S + +LA S+ + D + N +R
Sbjct: 260 PKSKNLVIPEEQSKSVRLEEELAGGSVCVGDLENNNEGRR 379
>TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial (7%)
Length = 841
Score = 28.9 bits (63), Expect = 0.33
Identities = 13/29 (44%), Positives = 16/29 (54%)
Frame = +2
Query: 83 PKPITVSSLTHGSSPTPYSILRVFHNSSS 111
P P T S+L H SP P + + HN SS
Sbjct: 125 PNPNTFSNLNHNLSPNPNTFSNLNHNLSS 211
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,923,672
Number of Sequences: 28460
Number of extensions: 53545
Number of successful extensions: 314
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 308
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 312
length of query: 138
length of database: 4,897,600
effective HSP length: 81
effective length of query: 57
effective length of database: 2,592,340
effective search space: 147763380
effective search space used: 147763380
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 50 (23.9 bits)
Lotus: description of TM0019b.9


