

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0019a.6
(1380 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
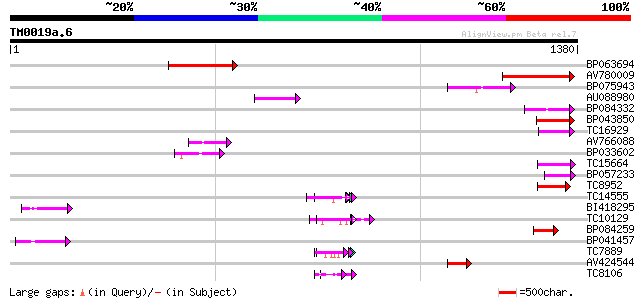
Score E
Sequences producing significant alignments: (bits) Value
BP063694 290 8e-79
AV780009 155 5e-38
BP075943 100 2e-21
AU088980 84 1e-16
BP084332 79 4e-15
BP043850 79 4e-15
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 75 9e-14
AV766088 69 4e-12
BP033602 68 9e-12
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 68 9e-12
BP057233 66 3e-11
TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%) 66 4e-11
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 59 4e-09
BI418295 59 4e-09
TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%) 59 5e-09
BP084259 58 1e-08
BP041457 55 7e-08
TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall prot... 55 1e-07
AV424544 54 2e-07
TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cu... 53 4e-07
>BP063694
Length = 511
Score = 290 bits (743), Expect = 8e-79
Identities = 140/169 (82%), Positives = 154/169 (90%)
Frame = -2
Query: 386 IFEADSFVIQNRQTGAVLGKGPCDKGFYVLDQGSQALLATSSSLPRASFELWHSRLDHVN 445
IF DSFVIQNR+TGAVLG+ CD+G YVL+Q SQALL TSS LPRASFELWHSRL HVN
Sbjct: 510 IFLDDSFVIQNRKTGAVLGRXRCDQGLYVLNQDSQALLTTSSPLPRASFELWHSRLGHVN 331
Query: 446 FDIIKKLHKLGCFNVSSILPKPICCTSCQMAKSKRLVCYDNHKRASAVLDLVHCDLWGPS 505
FDIIK+LHK GC +VSSILPKPICCTSCQMAKSKRLV +DN+KRASAVLDL+HCDL GPS
Sbjct: 330 FDIIKQLHKHGCLDVSSILPKPICCTSCQMAKSKRLVFHDNNKRASAVLDLIHCDLRGPS 151
Query: 506 PVASVDGFSYFVIFVDDFSQFTWFYPLKRKSDFYDVLVRFKAFVENQFS 554
PVAS+DGFSYFVIFVDDFS+FTWFYPLKRKSDF DVL+RFK F+EN+FS
Sbjct: 150 PVASIDGFSYFVIFVDDFSRFTWFYPLKRKSDFSDVLLRFKVFMENRFS 4
>AV780009
Length = 529
Score = 155 bits (391), Expect = 5e-38
Identities = 81/176 (46%), Positives = 106/176 (60%)
Frame = -1
Query: 1199 QAVKRIIRYVCAMQHFGLTFRRTSSPAVLGYSDADWARCTDHRRSTYGYAIFLGYNLLSW 1258
QA R++RYV GL F S + YSD+DWA C D RRS GY+IFLG +L+SW
Sbjct: 529 QAATRVLRYVKGAPAQGLFFSADSPLKLQAYSDSDWAGCPDTRRSVTGYSIFLGTSLISW 350
Query: 1259 SAKKQPSVAHSSCESEYRAMANTASELVWLLNLLHELRVRLSAPPLFLSDNQSALFMAQN 1318
KKQ +V+ SS E+EYRA+A T E+ WL L L++ + P DNQSAL +A N
Sbjct: 349 RTKKQTTVSRSSSEAEYRALAATVCEVQWLSYLFQFLKLNVPLPVPLFCDNQSALHIAHN 170
Query: 1319 PVAHKHAKHIDLDCHFVRELVSSGRLAVRHVPTSLQLADIFTKVLPRPLFDLFRSK 1374
P H+ KHI+LDCH VR + +G + + + T QLADIFTK P F L ++
Sbjct: 169 PTFHERTKHIELDCHVVRAKLQAGLIHLLPISTHHQLADIFTKPCPHLCFPLLGAR 2
>BP075943
Length = 547
Score = 100 bits (249), Expect = 2e-21
Identities = 65/176 (36%), Positives = 96/176 (53%), Gaps = 9/176 (5%)
Frame = -3
Query: 1065 LLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYLGKLGYFLGLEITYTADGLFLGHAKYA 1124
LL+YVDD++L G+D + + L+D+F IK LG+ YFLGLEI + G+ L KYA
Sbjct: 521 LLLYVDDVLLAGNDMHEIQLVKSSLHDQFRIKDLGEAKYFLGLEIARSTSGIVLNQRKYA 342
Query: 1125 HDLLSHALMLE-----ASHVLTPLAAGSHLVSSGEGYSDPTHYRSLVGALQYLTITRPDL 1179
L+S + SH G+ ++G +D YR +VG L YL TRPD+
Sbjct: 341 LQLISDSGHFGFQPRFYSHGTNSQTLGT---NTGTPLTDIGSYRRIVGRLLYLNTTRPDI 171
Query: 1180 SYAVNTVSQFLQTPTVDHFQAVKRIIRYVCAMQHFGLTFRRTSS----PAVLGYSD 1231
++AVN +SQFL PT H Q + ++Y+ GL + +SS P+++G D
Sbjct: 170 TFAVNQLSQFLSAPTDIHEQQLTGFLKYL*GSPGSGLFYPASSSTL*LPSLIGGPD 3
>AU088980
Length = 360
Score = 84.3 bits (207), Expect = 1e-16
Identities = 41/113 (36%), Positives = 61/113 (53%), Gaps = 1/113 (0%)
Frame = +2
Query: 595 NGRVERKHRHVLELGLAMLYHSHVSTSYWVHAFSTAVYIINRVPSKVLSDQIPFQLLFQV 654
N VERKHRH+L + A+++HS++ + W A A Y+IN +PS +L Q P+Q L
Sbjct: 2 NSIVERKHRHILNVTRALMFHSYLPKNLWTFAVKHAAYLINXLPSPLLKGQCPYQFLNND 181
Query: 655 APTYANFHPFGCRVFPCLRPYMKNKFSPRGTPCIFLGYNSHHKGFKCFD-PTT 706
+PT + FG + + K PR C+FLG+ + G+ D PTT
Sbjct: 182 SPTLLDLKVFGTLCYSSTLTHNXQKLDPRARKCVFLGFKTGTXGYIVMDLPTT 340
>BP084332
Length = 368
Score = 79.3 bits (194), Expect = 4e-15
Identities = 42/123 (34%), Positives = 73/123 (59%), Gaps = 1/123 (0%)
Frame = +2
Query: 1254 NLLSWSAK-KQPSVAHSSCESEYRAMANTASELVWLLNLLHELRVRLSAPPLFLSDNQSA 1312
N+ W A Q ++A S+ E+EY + A +++++W+ + L + ++ S P++ DN +A
Sbjct: 2 NVNFWEAT*GQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQILESNIPIYC-DNTAA 178
Query: 1313 LFMAQNPVAHKHAKHIDLDCHFVRELVSSGRLAVRHVPTSLQLADIFTKVLPRPLFDLFR 1372
+ +++NP+ H AKHI++ HF+R+ V G L ++ V T Q ADIFTK L F+
Sbjct: 179 ISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFIL 358
Query: 1373 SKL 1375
L
Sbjct: 359 KNL 367
>BP043850
Length = 515
Score = 79.3 bits (194), Expect = 4e-15
Identities = 42/93 (45%), Positives = 56/93 (60%)
Frame = -1
Query: 1283 SELVWLLNLLHELRVRLSAPPLFLSDNQSALFMAQNPVAHKHAKHIDLDCHFVRELVSSG 1342
SE++WL LL EL S P +DN SA+ +A NPV H+ +HI++DCH VRE
Sbjct: 515 SEIIWLRGLLSELGFLQSQPTPLHADNTSAIQIAANPVYHEWTRHIEVDCHSVREAYDRR 336
Query: 1343 RLAVRHVPTSLQLADIFTKVLPRPLFDLFRSKL 1375
+ + HV TS+Q+ADI TK L R + SKL
Sbjct: 335 VITLPHVSTSVQIADILTKSLTRQRHNFLVSKL 237
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 74.7 bits (182), Expect = 9e-14
Identities = 38/89 (42%), Positives = 51/89 (56%)
Frame = +1
Query: 1287 WLLNLLHELRVRLSAPPLFLSDNQSALFMAQNPVAHKHAKHIDLDCHFVRELVSSGRLAV 1346
WL LL +L+V P L DN SA +A NPV H+ KHI++DCH VRE + G + +
Sbjct: 4 WLTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDCHIVRERIQKGLIHL 183
Query: 1347 RHVPTSLQLADIFTKVLPRPLFDLFRSKL 1375
+ +S LADI+TK L F +KL
Sbjct: 184 LPISSSEPLADIYTKALSPQNFHQICAKL 270
>AV766088
Length = 501
Score = 69.3 bits (168), Expect = 4e-12
Identities = 36/104 (34%), Positives = 53/104 (50%)
Frame = -3
Query: 436 LWHSRLDHVNFDIIKKLHKLGCFNVSSILPKPICCTSCQMAKSKRLVCYDNHKRASAVLD 495
LWHSRL H + ++ L + + P+C SC K RL ++ D
Sbjct: 307 LWHSRLGHPSSSALRYLRSNKFISYELLNYSPVC-ESCVFGKHVRLPFVSSNNVTVMPFD 131
Query: 496 LVHCDLWGPSPVASVDGFSYFVIFVDDFSQFTWFYPLKRKSDFY 539
++H DLW SPV S G ++V+F+DDF+ F W +PL KS +
Sbjct: 130 ILHSDLW-TSPVLSSAGHRFYVLFLDDFTDFLWTFPLSNKSQVF 2
>BP033602
Length = 533
Score = 68.2 bits (165), Expect = 9e-12
Identities = 40/126 (31%), Positives = 60/126 (46%), Gaps = 5/126 (3%)
Frame = +3
Query: 402 VLGKGPCDKGFYVLD-----QGSQALLATSSSLPRASFELWHSRLDHVNFDIIKKLHKLG 456
++G G Y + + +Q L S + R LWH+RL H NF ++ L
Sbjct: 159 MIGSAKMKDGLYYFEDTFRNKAAQGLCGMSCTSVRDQIMLWHNRLGHPNFQYLRHLFPDL 338
Query: 457 CFNVSSILPKPICCTSCQMAKSKRLVCYDNHKRASAVLDLVHCDLWGPSPVASVDGFSYF 516
NV+ + C SC +AK++R Y AS L+H D+WGPS + + G +F
Sbjct: 339 FKNVNC---SSLECESCVLAKNQRAPYYSQPYHASRPFYLIHSDVWGPSKITTQFGKRWF 509
Query: 517 VIFVDD 522
V F+DD
Sbjct: 510 VTFIDD 527
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 68.2 bits (165), Expect = 9e-12
Identities = 39/94 (41%), Positives = 52/94 (54%)
Frame = +1
Query: 1284 ELVWLLNLLHELRVRLSAPPLFLSDNQSALFMAQNPVAHKHAKHIDLDCHFVRELVSSGR 1343
EL WL +L +LRV +P L D+QSA +A N V H+ KH+D+DCH VRE + +
Sbjct: 4 EL*WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDCHVVREKLQAKL 183
Query: 1344 LAVRHVPTSLQLADIFTKVLPRPLFDLFRSKLRV 1377
+ + + Q ADI TK L F SKL V
Sbjct: 184 FHLLPISSVDQTADILTKPLESGPFSHLVSKLGV 285
>BP057233
Length = 473
Score = 66.2 bits (160), Expect = 3e-11
Identities = 32/75 (42%), Positives = 44/75 (58%)
Frame = -2
Query: 1303 PLFLSDNQSALFMAQNPVAHKHAKHIDLDCHFVRELVSSGRLAVRHVPTSLQLADIFTKV 1362
P+ DN SA +A NPV H +KHI++D H++R+ V + V +VPT+ Q+AD TK
Sbjct: 451 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNEVVVAYVPTTDQIADCLTKP 272
Query: 1363 LPRPLFDLFRSKLRV 1377
L F R KL V
Sbjct: 271 LSHTRFSQLRDKLGV 227
>TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%)
Length = 550
Score = 65.9 bits (159), Expect = 4e-11
Identities = 34/80 (42%), Positives = 49/80 (60%)
Frame = +3
Query: 1284 ELVWLLNLLHELRVRLSAPPLFLSDNQSALFMAQNPVAHKHAKHIDLDCHFVRELVSSGR 1343
E +WL L +LR+ + DN+SAL +A N V HK ++I++DCH V V G
Sbjct: 21 EALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFGI 200
Query: 1344 LAVRHVPTSLQLADIFTKVL 1363
L + HVP+S Q+AD+FTK +
Sbjct: 201 LHLLHVPSSDQVADVFTKTI 260
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 59.3 bits (142), Expect = 4e-09
Identities = 47/127 (37%), Positives = 55/127 (43%), Gaps = 5/127 (3%)
Frame = +2
Query: 723 PLTGSKSSPNDLVVFTFYEPAA-GSPPSLQPVILVVPESSPPTG-PLPCPSCVDPDVQPV 780
P +SP T PAA SPP Q V S PP P P S P P
Sbjct: 191 PSNSPTTSPAPPTPTTPQPPAAQSSPPPAQSSPPPVQSSPPPASTPPPAQSSPPPVSSPP 370
Query: 781 PVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPL---APVD 837
PV PP P P + PP P PPATP PP + P PPA+T VP AP
Sbjct: 371 PVQSTPP-PAPASTPPPASPPPFSPPPATP-----PPPAATP--PPALTPVPATSPAPAP 526
Query: 838 ARPRTRS 844
A+ +++S
Sbjct: 527 AKVKSKS 547
Score = 48.1 bits (113), Expect = 9e-06
Identities = 28/88 (31%), Positives = 38/88 (42%)
Frame = +2
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
PA+ PP+ P P S PP P P + P + PVP P+P P PAP
Sbjct: 398 PASTPPPASPP-----PFSPPPATPPPPAATPPPALTPVPATSPAPAPAKVKSKSPAPAP 562
Query: 802 TSPTPPATPLVVTQAPPTSPPTTPPAVT 829
P L ++AP S + PA++
Sbjct: 563 APALAPVVSLSPSEAPGPSLSSLSPALS 646
Score = 43.5 bits (101), Expect = 2e-04
Identities = 38/108 (35%), Positives = 42/108 (38%), Gaps = 11/108 (10%)
Frame = +2
Query: 742 PAAGSPPSLQ----PVILVVP-ESSPPTGPLPCPSCVDPDVQPVPVDDAPP------SPV 790
PA SPP +Q P P +SSPP P+ P V P P PP SP
Sbjct: 272 PAQSSPPPVQSSPPPASTPPPAQSSPP--PVSSPPPVQSTPPPAPASTPPPASPPPFSPP 445
Query: 791 PHNDAPPLPAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDA 838
P PP P PPA +T P TSP P V AP A
Sbjct: 446 PATPPPPAATP----PPA----LTPVPATSPAPAPAKVKSKSPAPAPA 565
Score = 43.1 bits (100), Expect = 3e-04
Identities = 31/93 (33%), Positives = 37/93 (39%), Gaps = 2/93 (2%)
Frame = +2
Query: 742 PAAGSPPSLQPVILVVP-ESSPPTGPLPCPSCVDPD-VQPVPVDDAPPSPVPHNDAPPLP 799
PA SPP PV P +S+PP P P P P P PP+ P P+P
Sbjct: 332 PAQSSPP---PVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAATPPPALTPVP 502
Query: 800 APTSPTPPATPLVVTQAPPTSPPTTPPAVTQVP 832
A TSP P + P P P V+ P
Sbjct: 503 A-TSPAPAPAKVKSKSPAPAPAPALAPVVSLSP 598
Score = 31.6 bits (70), Expect = 0.89
Identities = 25/69 (36%), Positives = 35/69 (50%), Gaps = 9/69 (13%)
Frame = +2
Query: 785 APPSPVPHNDAPPLPAPTSPTPPA---TPLVVTQAPP---TSPP---TTPPAVTQVPLAP 835
+P + + APP P T+P PPA +P +PP +SPP T PPA + P P
Sbjct: 188 SPSNSPTTSPAPPTP--TTPQPPAAQSSPPPAQSSPPPVQSSPPPASTPPPAQSSPP--P 355
Query: 836 VDARPRTRS 844
V + P +S
Sbjct: 356 VSSPPPVQS 382
>BI418295
Length = 523
Score = 59.3 bits (142), Expect = 4e-09
Identities = 35/125 (28%), Positives = 65/125 (52%), Gaps = 1/125 (0%)
Frame = +2
Query: 29 WSKQVTAILQNQNLFD-IVDPIVAPPSEFMLQPVSSALHHVVPAPNPLFVQWQARDRNVN 87
W +QV +Q+ L D +VDP + P F+ + V NP + W +D V
Sbjct: 14 WRQQVEGAIQSHQLHDFLVDPEI--PQRFLTEE-----DRVAATENPAYTTWIKQDSMVF 172
Query: 88 SLLLSSLTEEALSETLSATTARDVWTALHSAYASRNKAREMRLRDELQLMRRGSLSVSEY 147
+ LL++L+ L ++ + VW + + ++ +A+ +LR EL+ + +GS S++EY
Sbjct: 173 TWLLTTLSPFVLPTVINCERSSQVWREILDFFENQCRAQSAKLRSELKNITKGSKSITEY 352
Query: 148 GRKIR 152
R+I+
Sbjct: 353 LRRIK 367
>TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%)
Length = 562
Score = 58.9 bits (141), Expect = 5e-09
Identities = 43/112 (38%), Positives = 55/112 (48%), Gaps = 13/112 (11%)
Frame = +1
Query: 746 SPPSL-QPVILVVPESSPPTGPLPCPSCVDPDVQP--VPVDDAPPSPV---PHNDAPP-L 798
SPP++ P P + PP P P P P QP +P+ PSP+ PH PP
Sbjct: 112 SPPNVPSPPKAPSPNNHPPYTPTP-PKTPSPTSQPPYIPLPPKTPSPISQPPHVPTPPST 288
Query: 799 PAPTS-----PTPPATPLVVTQAPPT-SPPTTPPAVTQVPLAPVDARPRTRS 844
P+P S PTPP TP +Q P T +PP TP ++Q P P P+T S
Sbjct: 289 PSPISQPPYTPTPPKTPSPTSQPPHTPTPPNTPSPISQPPYTPTP--PKTPS 438
Score = 56.6 bits (135), Expect = 3e-08
Identities = 45/153 (29%), Positives = 67/153 (43%), Gaps = 12/153 (7%)
Frame = +1
Query: 746 SPPSLQ-PVILVVPESSPPTGPLP----CPSCVDPDVQPVPVDDAPPSPVPHNDAPP-LP 799
+PPS+ P P + PP P+P PS P+V P +P + P+ PP P
Sbjct: 16 TPPSIPTPPKTPSPGNQPPHTPIPPKTPSPSYSPPNVPSPPKAPSPNNHPPYTPTPPKTP 195
Query: 800 APTS-----PTPPATPLVVTQAPPT-SPPTTPPAVTQVPLAPVDARPRTRS*NGIFKPNP 853
+PTS P PP TP ++Q P +PP+TP ++Q P P + + + P P
Sbjct: 196 SPTSQPPYIPLPPKTPSPISQPPHVPTPPSTPSPISQPPYTPTPPKTPSPTSQPPHTPTP 375
Query: 854 RYALVHAQPTGLLTALHTVT*PKGFKSAMKHPH 886
P+ + +T T PK + PH
Sbjct: 376 PNT-----PSPISQPPYTPTPPKTPSPTNQPPH 459
Score = 55.8 bits (133), Expect = 4e-08
Identities = 49/139 (35%), Positives = 57/139 (40%), Gaps = 26/139 (18%)
Frame = +1
Query: 730 SPNDLVVFTFYEPAAGSPPSLQPVILVVPE-----SSPPTGPLPCPSCVDPDVQP--VPV 782
SPN+ +T P SP S P I + P+ S PP P P PS P QP P
Sbjct: 148 SPNNHPPYTPTPPKTPSPTSQPPYIPLPPKTPSPISQPPHVPTP-PSTPSPISQPPYTPT 324
Query: 783 DDAPPSPV---PHNDAPP-LPAPTS-----PTPPATPLVVTQAP----------PTSPPT 823
PSP PH PP P+P S PTPP TP Q P PTS P
Sbjct: 325 PPKTPSPTSQPPHTPTPPNTPSPISQPPYTPTPPKTPSPTNQPPHIPSPPNSSSPTSQPP 504
Query: 824 TPPAVTQVPLAPVDARPRT 842
P+ + P +P P T
Sbjct: 505 NTPSPPKTP-SPTSQPPNT 558
>BP084259
Length = 385
Score = 57.8 bits (138), Expect = 1e-08
Identities = 26/59 (44%), Positives = 36/59 (60%)
Frame = -3
Query: 1276 RAMANTASELVWLLNLLHELRVRLSAPPLFLSDNQSALFMAQNPVAHKHAKHIDLDCHF 1334
R + +TA+E +WL LL EL+ L P S+N +A ++ N V H H KH+ LDCHF
Sbjct: 179 RTIGSTAAEFLWLQQLLKELQFSLPKVPCIFSNNINATYVCLNLVFHSHMKHVALDCHF 3
>BP041457
Length = 495
Score = 55.1 bits (131), Expect = 7e-08
Identities = 35/134 (26%), Positives = 65/134 (48%), Gaps = 1/134 (0%)
Frame = +2
Query: 15 HMITVKLNSGNFLLWSKQVTAILQNQNLFD-IVDPIVAPPSEFMLQPVSSALHHVVPAPN 73
++I KL++ FLLW +QV I++ +L IV+P + P SS H N
Sbjct: 107 YLIFEKLSAKKFLLWKQQVEPIIKAHHLHCYIVEPAIPPR-------YSSDEEHANGVVN 265
Query: 74 PLFVQWQARDRNVNSLLLSSLTEEALSETLSATTARDVWTALHSAYASRNKAREMRLRDE 133
P + W+ D+ + S L S L+ + + + +W +HS + ++ +A+ ++R E
Sbjct: 266 PAYPIWETHDQFLLSWLQSLLSSSVHTCMIGCVNSFQLWVKIHSFFHNQTRAKSTQIRTE 445
Query: 134 LQLMRRGSLSVSEY 147
L+ + G + Y
Sbjct: 446 LRSAKLGGRLLHGY 487
>TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (66%)
Length = 1158
Score = 54.7 bits (130), Expect = 1e-07
Identities = 37/104 (35%), Positives = 44/104 (41%), Gaps = 8/104 (7%)
Frame = +1
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPL-------PCPSCVDPDVQPVPVDDAPPSPVPHND 794
P PP ++P I V P PT P+ P P P V P P PP+P P
Sbjct: 247 PYVPKPPIVKPPI-VYPPPYVPTPPIVKPPIVYPPPYVPLPPVVPSPPIVTPPTPTPPIV 423
Query: 795 APPLPAPTSPTPPATPLVVTQAP-PTSPPTTPPAVTQVPLAPVD 837
PP P P TPP+ P T+ P P PP P P P+D
Sbjct: 424 TPPTPTPPVVTPPSPP---TETPCPPPPPVVPSPPPAQPTCPID 546
Score = 50.4 bits (119), Expect = 2e-06
Identities = 30/85 (35%), Positives = 39/85 (45%), Gaps = 2/85 (2%)
Frame = +1
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P +PP ++P I+ P P +P P V P P P PP+P P PP P
Sbjct: 298 PYVPTPPIVKPPIVYPPPYVPLPPVVPSPPIVTPPT-PTPPIVTPPTPTPPVVTPPSPPT 474
Query: 802 TSPTPPATPLVVTQAP--PTSPPTT 824
+P PP P+V + P PT P T
Sbjct: 475 ETPCPPPPPVVPSPPPAQPTCPIDT 549
Score = 45.1 bits (105), Expect = 8e-05
Identities = 38/116 (32%), Positives = 47/116 (39%), Gaps = 21/116 (18%)
Frame = +1
Query: 741 EPAAGSPPSLQ--PVILVVPE--SSPPTGPLPCPSCVDPD-VQPVPVDDAPPS------- 788
+P SPP + PV+ V P PP P+ P P V P P PP
Sbjct: 112 KPPVVSPPYVPKPPVVPVTPPYVPKPPVVPVTPPYVPKPPIVYPPPYVPKPPIVKPPIVY 291
Query: 789 PVPHNDAPPL-------PAPTSPTPPA--TPLVVTQAPPTSPPTTPPAVTQVPLAP 835
P P+ PP+ P P P PP +P +VT PT P TPP T + P
Sbjct: 292 PPPYVPTPPIVKPPIVYPPPYVPLPPVVPSPPIVTPPTPTPPIVTPPTPTPPVVTP 459
Score = 42.7 bits (99), Expect = 4e-04
Identities = 33/95 (34%), Positives = 39/95 (40%), Gaps = 3/95 (3%)
Frame = +1
Query: 746 SPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPT 805
+PP + +V P PP P P P P V P P PP P PP P P
Sbjct: 205 TPPYVPKPPIVYP---PPYVPKP-PIVKPPIVYPPPYVPTPPIVKPPIVYPPPYVPLPPV 372
Query: 806 PPATPLVV--TQAPPTS-PPTTPPAVTQVPLAPVD 837
P+ P+V T PP PPT P V P P +
Sbjct: 373 VPSPPIVTPPTPTPPIVTPPTPTPPVVTPPSPPTE 477
Score = 42.0 bits (97), Expect = 7e-04
Identities = 39/118 (33%), Positives = 46/118 (38%), Gaps = 19/118 (16%)
Frame = +1
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVP---------VDDAPPSPV-- 790
P PP ++P + P P +P P V P P P V P PV
Sbjct: 31 PHVPKPPIVKPPYVPKPPVVKPPPYVPKPPVVSPPYVPKPPVVPVTPPYVPKPPVVPVTP 210
Query: 791 PHNDAPPL--PAPTSPTPPAT-PLVVTQAP--PTSPPTTPPAV---TQVPLAPVDARP 840
P+ PP+ P P P PP P +V P PT P PP V VPL PV P
Sbjct: 211 PYVPKPPIVYPPPYVPKPPIVKPPIVYPPPYVPTPPIVKPPIVYPPPYVPLPPVVPSP 384
Score = 40.4 bits (93), Expect = 0.002
Identities = 26/70 (37%), Positives = 28/70 (39%)
Frame = +1
Query: 741 EPAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPA 800
+P PP P+ VVP T P P P V P PV P P PP P
Sbjct: 325 KPPIVYPPPYVPLPPVVPSPPIVTPPTPTPPIVTPPTPTPPVVTPPSPPTETPCPPPPPV 504
Query: 801 PTSPTPPATP 810
SP PPA P
Sbjct: 505 VPSP-PPAQP 531
Score = 39.3 bits (90), Expect = 0.004
Identities = 33/110 (30%), Positives = 42/110 (38%), Gaps = 11/110 (10%)
Frame = +1
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPV----PHNDAPP 797
P PP + P VP+ PP P+ P P V PV P P+ P+ PP
Sbjct: 100 PYVPKPPVVSPPY--VPK--PPVVPVTPPYVPKPPVVPVTPPYVPKPPIVYPPPYVPKPP 267
Query: 798 L-------PAPTSPTPPATPLVVTQAPPTSPPTTPPAVTQVPLAPVDARP 840
+ P P PTPP + + P PP P VP P+ P
Sbjct: 268 IVKPPIVYPPPYVPTPP-----IVKPPIVYPPPYVPLPPVVPSPPIVTPP 402
Score = 38.1 bits (87), Expect = 0.009
Identities = 25/70 (35%), Positives = 27/70 (37%)
Frame = +1
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P SPP + P P +PPT P P V P P PP PV P P P
Sbjct: 367 PVVPSPPIVTPPTPTPPIVTPPT---PTPPVVTPPSPPTETPCPPPPPV-----VPSPPP 522
Query: 802 TSPTPPATPL 811
PT P L
Sbjct: 523 AQPTCPIDTL 552
>AV424544
Length = 276
Score = 53.9 bits (128), Expect = 2e-07
Identities = 29/59 (49%), Positives = 39/59 (65%)
Frame = +3
Query: 1065 LLVYVDDIILTGSDPSLLT*FIACLNDEFAIKYLGKLGYFLGLEITYTADGLFLGHAKY 1123
+LVYVDD+IL G+D + + L+ +F IK LG L YFLGLE+ ++ GLFL KY
Sbjct: 99 ILVYVDDLILAGNDLNEIQCVKNKLDIQFRIKDLGTLKYFLGLEVARSSCGLFLSQRKY 275
>TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber
hypocotyls (Arabinogalactan protein), partial (32%)
Length = 760
Score = 52.8 bits (125), Expect = 4e-07
Identities = 32/79 (40%), Positives = 38/79 (47%)
Frame = +1
Query: 742 PAAGSPPSLQPVILVVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAP 801
P A +P S P VP SSPP P+P S PP+PVP + +PP PAP
Sbjct: 76 PVAKAPASTPPA--AVPVSSPPPAPVPVAS--------------PPAPVPVS-SPPAPAP 204
Query: 802 TSPTPPATPLVVTQAPPTS 820
T+P P TP AP S
Sbjct: 205 TTPAPAVTPTAEVPAPAPS 261
Score = 47.4 bits (111), Expect = 2e-05
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 1/89 (1%)
Frame = +1
Query: 756 VVPESSPPTGPLPCPSCVDPDVQPVPVDDAPPSPVPHNDAPPLPAPTSPTPPATPLVVTQ 815
V P S+P P + V P PP+ VP + PP P P + P P+
Sbjct: 16 VTPVSAPLASPPSKAAAAPAPVAKAPAS-TPPAAVPVSSPPPAPVPVASPPAPVPV---S 183
Query: 816 APPTSPPTTP-PAVTQVPLAPVDARPRTR 843
+PP PTTP PAVT P A +++
Sbjct: 184 SPPAPAPTTPAPAVTPTAEVPAPAPSKSK 270
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.138 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,999,665
Number of Sequences: 28460
Number of extensions: 516632
Number of successful extensions: 12997
Number of sequences better than 10.0: 841
Number of HSP's better than 10.0 without gapping: 5824
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8871
length of query: 1380
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1278
effective length of database: 1,994,680
effective search space: 2549201040
effective search space used: 2549201040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0019a.6


