

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
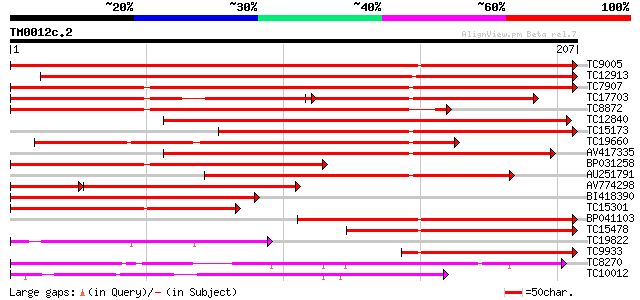
Query= TM0012c.2
(207 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9005 similar to UP|Q84L76 (Q84L76) Expansin precursor, partial... 293 1e-80
TC12913 homologue to UP|Q84L76 (Q84L76) Expansin precursor, part... 284 9e-78
TC7907 homologue to UP|O81133 (O81133) Expansin, partial (97%) 270 1e-73
TC17703 homologue to UP|CAC19183 (CAC19183) Expansin, partial (82%) 141 3e-56
TC8872 homologue to UP|Q8L5G5 (Q8L5G5) Alpha-expansin 3, partial... 204 7e-54
TC12840 similar to PIR|T02010|T02010 expansin homolog T15B16.16 ... 188 5e-49
TC15173 similar to UP|AAR09168 (AAR09168) Alpha-expansin 1, part... 177 2e-45
TC19660 similar to GB|BAB09383.1|9758857|AB010694 expansin-like ... 176 2e-45
AV417335 174 1e-44
BP031258 169 2e-43
AU251791 155 4e-39
AV774298 134 2e-37
BI418390 145 6e-36
TC15301 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 prec... 114 2e-26
BP041103 107 1e-24
TC15478 homologue to UP|AAQ08016 (AAQ08016) Expansin, partial (33%) 105 6e-24
TC19822 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 prec... 99 7e-22
TC9933 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 precu... 76 4e-15
TC8270 similar to UP|Q7XHJ2 (Q7XHJ2) Expansin-like protein (Frag... 75 6e-15
TC10012 weakly similar to UP|EXR1_ARATH (O23547) Expansin-relate... 69 6e-13
>TC9005 similar to UP|Q84L76 (Q84L76) Expansin precursor, partial (92%)
Length = 1277
Score = 293 bits (751), Expect = 1e-80
Identities = 131/207 (63%), Positives = 157/207 (75%)
Frame = +1
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY ++ GYG++TAA+S+ LF G CGACFEIKC P+WC PG PSI VTATN
Sbjct: 196 GACGYGNLYSQGYGVNTAALSTALFNNGLSCGACFEIKCDQDPRWCNPGSPSIIVTATNF 375
Query: 61 CPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIRFTVT 120
CPPN AQ +D+GGWCNPPR HFDLA P +LKIA+Y+ GIVPV YRRVPC +KGGIRFT+
Sbjct: 376 CPPNLAQPSDDGGWCNPPRTHFDLAMPMFLKIAQYRAGIVPVTYRRVPCRKKGGIRFTIN 555
Query: 121 GNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTFRVRA 180
G YFNLV + NV GAGDI ++ VKG W + RNWG+ W++N+ LVG++L+FRV
Sbjct: 556 GFRYFNLVLISNVAGAGDIVRVSVKGTNS-GWNSMSRNWGQNWQSNSDLVGQALSFRVTG 732
Query: 181 SDGRYSTAWKVAPKDWQFGQTFEGKNF 207
SD R ST+W VAP WQFGQTF GKNF
Sbjct: 733 SDRRTSTSWNVAPSHWQFGQTFTGKNF 813
>TC12913 homologue to UP|Q84L76 (Q84L76) Expansin precursor, partial (78%)
Length = 606
Score = 284 bits (726), Expect = 9e-78
Identities = 125/196 (63%), Positives = 152/196 (76%)
Frame = +2
Query: 12 GYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNLCPPNYAQGNDN 71
GYG++TAA+S+ LF G CGACFE+KC P+WC PG PSI VTATN CPPN+AQ +DN
Sbjct: 20 GYGVNTAALSTALFNNGLSCGACFEMKCDQDPRWCNPGNPSILVTATNFCPPNFAQPSDN 199
Query: 72 GGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIRFTVTGNPYFNLVKVW 131
GGWCNPPR HFDLA P +LKIA+Y+ GIVPV YRRVPC + GGIRFT+ G YFNLV +
Sbjct: 200 GGWCNPPRPHFDLAMPMFLKIAQYRAGIVPVAYRRVPCRKSGGIRFTINGFRYFNLVLIT 379
Query: 132 NVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTFRVRASDGRYSTAWKV 191
NV GAGDI ++ VKG W + RNWG+ W++N++LVG++L+FRV SD R ST+W V
Sbjct: 380 NVAGAGDIVRVSVKGG-NTAWMSMSRNWGQNWQSNSVLVGQTLSFRVTGSDRRTSTSWNV 556
Query: 192 APKDWQFGQTFEGKNF 207
AP +WQFGQTF GKNF
Sbjct: 557 APANWQFGQTFTGKNF 604
>TC7907 homologue to UP|O81133 (O81133) Expansin, partial (97%)
Length = 1313
Score = 270 bits (691), Expect = 1e-73
Identities = 119/207 (57%), Positives = 152/207 (72%)
Frame = +2
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY ++ GYG +TAA+S+ LF G CG+C+E++C + P+WC+PG SI VTATN
Sbjct: 227 GACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEMRCDSDPRWCLPG--SILVTATNF 400
Query: 61 CPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIRFTVT 120
CPPNYA N NGGWCNPP +HFDLA+PA+LKIA+YK GIVP+ +RRV C +KGGIRFT+
Sbjct: 401 CPPNYALSNSNGGWCNPPLQHFDLAEPAFLKIAQYKAGIVPISFRRVSCVKKGGIRFTIN 580
Query: 121 GNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTFRVRA 180
G+ YFNLV V NVGGAGD+ + +KG K W + RNWG+ W++N+ L G+SL+F+V
Sbjct: 581 GHSYFNLVLVTNVGGAGDVHSVSIKG-SKTGWQTMSRNWGQNWQSNSYLNGQSLSFKVTT 757
Query: 181 SDGRYSTAWKVAPKDWQFGQTFEGKNF 207
SDGR + P WQFGQTF G F
Sbjct: 758 SDGRTIISNNAVPAGWQFGQTFSGGQF 838
>TC17703 homologue to UP|CAC19183 (CAC19183) Expansin, partial (82%)
Length = 687
Score = 141 bits (355), Expect(2) = 3e-56
Identities = 65/112 (58%), Positives = 79/112 (70%)
Frame = +1
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY +++ GYG +TAA+S+ LF G CGACF+IKCVN PQWC+PG SI VTATN+
Sbjct: 139 GACGYGNLISQGYGTNTAALSTALFNSGLSCGACFQIKCVNDPQWCLPG--SIVVTATNV 312
Query: 61 CPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARK 112
CPP GG C+PP HFDL++P + IA+YK GIVPV YRRV K
Sbjct: 313 CPP--------GGGCDPPNHHFDLSQPIFQHIAQYKAGIVPVAYRRVQMQEK 444
Score = 93.2 bits (230), Expect(2) = 3e-56
Identities = 42/85 (49%), Positives = 59/85 (69%)
Frame = +2
Query: 109 CARKGGIRFTVTGNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAM 168
C RK GIRFT+ G+ YFNLV V NVGGAGD+ + +KG + +W + RNWG+ W++N+
Sbjct: 434 CRRKDGIRFTINGHSYFNLVLVTNVGGAGDVHAVAIKG-SRTRWQAMSRNWGQNWQSNSY 610
Query: 169 LVGESLTFRVRASDGRYSTAWKVAP 193
L G+SL+F V S+G ++ VAP
Sbjct: 611 LNGQSLSFVVTTSNGHSVVSFNVAP 685
>TC8872 homologue to UP|Q8L5G5 (Q8L5G5) Alpha-expansin 3, partial (82%)
Length = 729
Score = 204 bits (520), Expect = 7e-54
Identities = 92/161 (57%), Positives = 115/161 (71%)
Frame = +2
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY ++ GYG +TAA+S+ +F G CG+C+E++CVN +WC+PG SI VTATN
Sbjct: 209 GACGYGNLYSQGYGTNTAALSTAMFNNGLSCGSCYEVRCVNDHRWCLPG--SIVVTATNF 382
Query: 61 CPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIRFTVT 120
CPPN A N+ GGWCNPP HFDLA+P +L+IA+YK GIVPV YRRV C R+GGIRFT+
Sbjct: 383 CPPNNALPNNAGGWCNPPLHHFDLAQPVFLRIAQYKAGIVPVSYRRVACRRRGGIRFTIN 562
Query: 121 GNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGE 161
G+ YFNLV + NVGGAGD+ +KG RNW E
Sbjct: 563 GHSYFNLVLITNVGGAGDVHSAAIKG---------SRNWVE 658
Score = 35.8 bits (81), Expect = 0.005
Identities = 11/25 (44%), Positives = 19/25 (76%)
Frame = +3
Query: 152 WTDLKRNWGEKWETNAMLVGESLTF 176
W+ + NWG+ W++N+ L G+SL+F
Sbjct: 654 WSPMSTNWGQNWQSNSYLNGQSLSF 728
>TC12840 similar to PIR|T02010|T02010 expansin homolog T15B16.16 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(60%)
Length = 648
Score = 188 bits (478), Expect = 5e-49
Identities = 80/149 (53%), Positives = 108/149 (71%)
Frame = +3
Query: 57 ATNLCPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIR 116
ATN CPPN A NDNGGWCNPPR HFD+++PA+ IA+Y+ GIVP+ +RRV C + GGIR
Sbjct: 15 ATNFCPPNLALPNDNGGWCNPPRPHFDMSQPAFETIAKYRAGIVPIFHRRVRCRKSGGIR 194
Query: 117 FTVTGNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTF 176
FT+ G YF LV + NVGGAG+I+ + +KG K W + RNWG W++ + L G+SL+F
Sbjct: 195 FTINGRDYFELVLISNVGGAGEISMVWIKGSKMSSWETMSRNWGVNWQSLSYLNGQSLSF 374
Query: 177 RVRASDGRYSTAWKVAPKDWQFGQTFEGK 205
R++ +G+ TA+ VAP W+FGQ+F K
Sbjct: 375 RIQLRNGKTRTAYNVAPSSWKFGQSFTSK 461
>TC15173 similar to UP|AAR09168 (AAR09168) Alpha-expansin 1, partial (50%)
Length = 898
Score = 177 bits (448), Expect = 2e-45
Identities = 81/131 (61%), Positives = 98/131 (73%)
Frame = +2
Query: 77 PPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIRFTVTGNPYFNLVKVWNVGGA 136
PPR H DLA P +LKIAEY+ GIVPV YRRVPC + GGIRFT+ G YFNLV + NV GA
Sbjct: 2 PPRPHSDLAMPMFLKIAEYRSGIVPVAYRRVPCRKPGGIRFTINGFRYFNLVLISNVAGA 181
Query: 137 GDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTFRVRASDGRYSTAWKVAPKDW 196
GD+ + VKG + W + RNWG W++NA+LVG++L+FRV ASD R ST+W +AP W
Sbjct: 182 GDVVRTYVKG-TRTGWMPMSRNWGRNWQSNAVLVGQALSFRVTASDRRTSTSWNIAPAHW 358
Query: 197 QFGQTFEGKNF 207
QFGQTF GKNF
Sbjct: 359 QFGQTFTGKNF 391
>TC19660 similar to GB|BAB09383.1|9758857|AB010694 expansin-like protein
{Arabidopsis thaliana;} , partial (66%)
Length = 652
Score = 176 bits (447), Expect = 2e-45
Identities = 82/155 (52%), Positives = 104/155 (66%)
Frame = +2
Query: 10 KDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNLCPPNYAQGN 69
K GYGL T A+S+ LF G CGAC EIKCVNS WC PS+ VTATN CPPNY +
Sbjct: 197 KQGYGLHTTALSTALFNNGLACGACIEIKCVNSA-WCRKDAPSVLVTATNFCPPNYTK-- 367
Query: 70 DNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIRFTVTGNPYFNLVK 129
G CNPP++HFDL + KIA Y+ G+VPVQ RRVPC + GG++F + GNPYF +V
Sbjct: 368 TVGVRCNPPQKHFDLTVFMFTKIAWYRAGVVPVQARRVPCHKIGGVKFEIKGNPYFLMVL 547
Query: 130 VWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWE 164
V+NVG AGD++ + +KG W + NWG+ W+
Sbjct: 548 VYNVGNAGDVSSMSIKG-SNTGWLQMAHNWGQIWD 649
>AV417335
Length = 428
Score = 174 bits (440), Expect = 1e-44
Identities = 76/143 (53%), Positives = 99/143 (69%)
Frame = +3
Query: 57 ATNLCPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIR 116
ATN CPPN+A N+NGGWCNPP +HFD+A+PA+ KI Y+GGIVPV ++RVPC + GG+R
Sbjct: 3 ATNFCPPNFALPNNNGGWCNPPLKHFDMAQPAWEKIGIYRGGIVPVLFQRVPCKKHGGVR 182
Query: 117 FTVTGNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTF 176
F+V G YF LV + NV GAG I + +KG K W + RNWG W++NA L G+SL+F
Sbjct: 183 FSVNGRDYFELVMISNVAGAGSIQSVSIKG-SKTGWLSMSRNWGANWQSNAFLNGQSLSF 359
Query: 177 RVRASDGRYSTAWKVAPKDWQFG 199
R+ +DG V P +W FG
Sbjct: 360 RITTTDGETRVFPDVVPGNWAFG 428
>BP031258
Length = 526
Score = 169 bits (429), Expect = 2e-43
Identities = 72/116 (62%), Positives = 93/116 (80%)
Frame = +1
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY ++ GYG DTAA+S+ LF G CG+C+E++C + P+WC PG SI VTATN
Sbjct: 184 GACGYGNLYSQGYGTDTAALSTALFNNGLSCGSCYEMRCDDDPRWCKPG--SIIVTATNF 357
Query: 61 CPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIR 116
CPPN + N+NGGWCNPP +HFD+A+PA+L+IA+Y+ GIVPV +RRVPCA+KGGIR
Sbjct: 358 CPPNPSLANNNGGWCNPPLQHFDMAEPAFLQIAQYRAGIVPVAFRRVPCAKKGGIR 525
>AU251791
Length = 343
Score = 155 bits (393), Expect = 4e-39
Identities = 66/113 (58%), Positives = 88/113 (77%)
Frame = +1
Query: 72 GGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKGGIRFTVTGNPYFNLVKVW 131
GGWCNPP +HFDL++PA+ +IA+YK GIVPV YRRVPC +KGGI FT+ G+ YFNLV +
Sbjct: 4 GGWCNPPLQHFDLSQPAFQQIAQYKAGIVPVAYRRVPCQKKGGISFTINGHSYFNLVLIT 183
Query: 132 NVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTFRVRASDGR 184
NVGG+GD+ + +KG + W + RNWG+ W++NA L G+SL+F+V SDGR
Sbjct: 184 NVGGSGDVQAVSIKG-SRTDWQPMSRNWGQNWQSNANLNGQSLSFKVTTSDGR 339
>AV774298
Length = 482
Score = 134 bits (336), Expect(2) = 2e-37
Identities = 58/79 (73%), Positives = 61/79 (76%)
Frame = +2
Query: 28 GKICGACFEIKCVNSPQWCIPGQPSITVTATNLCPPNYAQGNDNGGWCNPPREHFDLAKP 87
G CGACFEIKC N QWC G PSI +TATN CPPNYA NDNGGWCNPPR FDLA P
Sbjct: 83 GLSCGACFEIKCANDKQWCHSGSPSIFITATNFCPPNYALPNDNGGWCNPPRPDFDLAMP 262
Query: 88 AYLKIAEYKGGIVPVQYRR 106
+LKIAEY+ GIVPV YRR
Sbjct: 263 MFLKIAEYRAGIVPVAYRR 319
Score = 37.7 bits (86), Expect(2) = 2e-37
Identities = 15/27 (55%), Positives = 21/27 (77%)
Frame = +1
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQE 27
GACGY ++ GYG++TAA+S+ LF E
Sbjct: 1 GACGYGNLYSQGYGVNTAALSTALFNE 81
>BI418390
Length = 471
Score = 145 bits (365), Expect = 6e-36
Identities = 60/91 (65%), Positives = 70/91 (75%)
Frame = +1
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY ++ GYG++TAA+S+ LF G CGACFE+KC N P WC G PSI VTATN
Sbjct: 199 GACGYGNLYSQGYGVNTAALSTALFNNGLSCGACFELKCANDPSWCHAGSPSIFVTATNF 378
Query: 61 CPPNYAQGNDNGGWCNPPREHFDLAKPAYLK 91
CPPN+AQ +DNGGWCNPPR HFDLA P +LK
Sbjct: 379 CPPNFAQASDNGGWCNPPRPHFDLAMPMFLK 471
>TC15301 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 precursor
(At-EXP13) (AtEx13) (Ath-ExpAlpha-1.22), partial (45%)
Length = 507
Score = 114 bits (284), Expect = 2e-26
Identities = 49/84 (58%), Positives = 60/84 (71%)
Frame = +1
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY D++K GYG+ TA +S LF+ G+ICGACFE++CV +WC+PG SI VTATN
Sbjct: 250 GACGYGDLLKTGYGMATAGLSETLFERGQICGACFELRCVEDSRWCLPG-TSIIVTATNF 426
Query: 61 CPPNYAQGNDNGGWCNPPREHFDL 84
C PNY + GG CNPP HF L
Sbjct: 427 CAPNYGFTAEGGGHCNPPNNHFVL 498
>BP041103
Length = 531
Score = 107 bits (268), Expect = 1e-24
Identities = 50/102 (49%), Positives = 66/102 (64%)
Frame = -1
Query: 106 RVPCARKGGIRFTVTGNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWET 165
RV C R+GGIRFT++G+ F V + N G GDI ++ KG + W + RNWG+ W
Sbjct: 531 RVKCRREGGIRFTISGSGIFISVLISNAAGMGDIVAVKTKGSRT-GWLPMGRNWGQNWHL 355
Query: 166 NAMLVGESLTFRVRASDGRYSTAWKVAPKDWQFGQTFEGKNF 207
NA+L + L+F V +SDG T + VAPK+W FGQTFEGK F
Sbjct: 354 NALLQNQPLSFEVTSSDGITLTCYNVAPKNWSFGQTFEGKQF 229
>TC15478 homologue to UP|AAQ08016 (AAQ08016) Expansin, partial (33%)
Length = 629
Score = 105 bits (262), Expect = 6e-24
Identities = 48/84 (57%), Positives = 60/84 (71%)
Frame = +1
Query: 124 YFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAMLVGESLTFRVRASDG 183
YFNLV + NV GAGDI + VKG W + RNWG+ W++N++LVG+SL+FRV ASD
Sbjct: 1 YFNLVLITNVAGAGDIVRTSVKGSDT-GWMSMSRNWGQNWQSNSVLVGQSLSFRVTASDR 177
Query: 184 RYSTAWKVAPKDWQFGQTFEGKNF 207
R ST+ + P WQFGQTF GKNF
Sbjct: 178 RTSTSLNIVPSHWQFGQTFTGKNF 249
>TC19822 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 precursor
(At-EXP13) (AtEx13) (Ath-ExpAlpha-1.22), partial (27%)
Length = 532
Score = 98.6 bits (244), Expect = 7e-22
Identities = 49/104 (47%), Positives = 62/104 (59%), Gaps = 8/104 (7%)
Frame = -1
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSP-------QWCIPGQPSI 53
GACGY + GYG TAA+S LF G+ICGACFE++CV +WC+ S+
Sbjct: 301 GACGYGN----GYGTATAALSQALFGRGQICGACFELRCVEEESATAFDRRWCVSSSTSV 134
Query: 54 TVTATNLCPPNYA-QGNDNGGWCNPPREHFDLAKPAYLKIAEYK 96
VTAT+ C PNY GG CNPP++HF L A+ KIA +K
Sbjct: 133 VVTATDFCAPNYGFDAESRGGRCNPPKQHFVLPVEAFEKIAIWK 2
>TC9933 similar to UP|EX13_ARATH (Q9M9P0) Alpha-expansin 13 precursor
(At-EXP13) (AtEx13) (Ath-ExpAlpha-1.22), partial (24%)
Length = 552
Score = 76.3 bits (186), Expect = 4e-15
Identities = 33/64 (51%), Positives = 42/64 (65%)
Frame = +1
Query: 144 VKGDKKLKWTDLKRNWGEKWETNAMLVGESLTFRVRASDGRYSTAWKVAPKDWQFGQTFE 203
VKG + W + RNWG+ W NA+L + L+F V SDG+ T++ APKDW FGQTFE
Sbjct: 1 VKGSRT-GWLQMGRNWGQNWHINALLQNQPLSFEVTGSDGKTVTSYNAAPKDWSFGQTFE 177
Query: 204 GKNF 207
GK F
Sbjct: 178 GKQF 189
>TC8270 similar to UP|Q7XHJ2 (Q7XHJ2) Expansin-like protein (Fragment),
partial (90%)
Length = 1006
Score = 75.5 bits (184), Expect = 6e-15
Identities = 63/217 (29%), Positives = 98/217 (45%), Gaps = 14/217 (6%)
Frame = +3
Query: 1 GACGYEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTATNL 60
GACGY+ + G AA LF+ G CGAC++++C N P C + V T+L
Sbjct: 153 GACGYDSLALGLSGGHLAAGVPSLFKSGAGCGACYQLRCKN-PTLC--KKEGARVVLTDL 323
Query: 61 CPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAE-------YKGGIVPVQYRRVPCARKG 113
P N + F L+ A++ +A+ K GIV ++Y+RV C K
Sbjct: 324 NPNNQTE--------------FVLSSRAFVAMAQDGKGQQILKLGIVDIEYKRVACEFKN 461
Query: 114 ---GIRFTVTG-NPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWGEKWETNAML 169
+R + P + +K GG +I + V W + RN G W+T+ +
Sbjct: 462 QNLAVRVEESSKKPDYLAIKFLYQGGQTEIVGVDVAQVGSSNWRFMSRNHGAVWDTSRVP 641
Query: 170 VGESLTFRVRAS---DGRYSTAWKVAPKDWQFGQTFE 203
G L FR+ + DG++ A KV P DW+ G ++
Sbjct: 642 QG-PLQFRLVVTAGYDGKWVWAQKVLPLDWKNGVIYD 749
>TC10012 weakly similar to UP|EXR1_ARATH (O23547) Expansin-related protein 1
precursor (At-EXPR1) (Ath-ExpBeta-3.1), partial (66%)
Length = 687
Score = 68.9 bits (167), Expect = 6e-13
Identities = 50/167 (29%), Positives = 78/167 (45%), Gaps = 7/167 (4%)
Frame = +2
Query: 1 GACG---YEDVVKDGYGLDTAAVSSVLFQEGKICGACFEIKCVNSPQWCIPGQPSITVTA 57
GACG Y +V DG A S L++ G CGAC++++C PQ+C + VT
Sbjct: 206 GACGFGEYGKIVNDG----NVAGVSWLWKNGSGCGACYQVRC-KIPQFCDDYGAYVVVTD 370
Query: 58 TNLCPPNYAQGNDNGGWCNPPREHFDLAKPAYLKIAEYKGGIVPVQYRRVPCARKG-GIR 116
+ D + PR + L K A +K G+V V+Y+RVPC G I
Sbjct: 371 FGV--------GDRTDFVMSPRGYSRLGKNADASAELFKYGVVDVEYKRVPCRYNGYNIL 526
Query: 117 FTV---TGNPYFNLVKVWNVGGAGDITQLQVKGDKKLKWTDLKRNWG 160
V + NP++ + + +GG DIT + + + W ++R +G
Sbjct: 527 LKVHERSNNPHYLAILIIYIGGTNDITAVDLWQEDCKDWRPMRRAFG 667
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.139 0.462
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,501,986
Number of Sequences: 28460
Number of extensions: 71785
Number of successful extensions: 394
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 370
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 370
length of query: 207
length of database: 4,897,600
effective HSP length: 86
effective length of query: 121
effective length of database: 2,450,040
effective search space: 296454840
effective search space used: 296454840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0012c.2


