

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
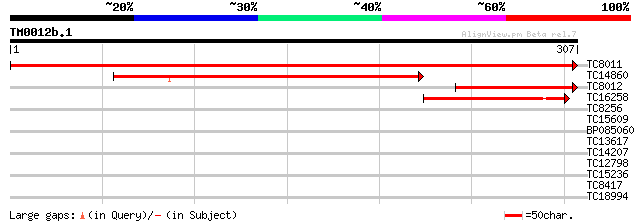
Query= TM0012b.1
(307 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8011 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomera... 605 e-174
TC14860 similar to UP|TPIS_COPJA (P21820) Triosephosphate isomer... 203 3e-53
TC8012 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomera... 118 1e-27
TC16258 similar to UP|TPIS_COPJA (P21820) Triosephosphate isomer... 110 3e-25
TC8256 homologue to UP|AAR12195 (AAR12195) Molecular chaperone H... 32 0.10
TC15609 homologue to UP|HS82_ARATH (P55737) Heat shock protein 8... 31 0.23
BP085060 28 2.0
TC13617 similar to UP|Q9FKY1 (Q9FKY1) Similarity to ribonucleopr... 28 2.5
TC14207 similar to UP|RK15_PEA (P31165) 50S ribosomal protein L1... 27 3.3
TC12798 similar to UP|O39876 (O39876) HBc antigen, partial (8%) 27 4.3
TC15236 homologue to UP|GCST_PEA (P49364) Aminomethyltransferase... 27 5.7
TC8417 similar to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimerase ... 26 9.7
TC18994 26 9.7
>TC8011 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomerase,
chloroplast precursor (TIM) , partial (89%)
Length = 1305
Score = 605 bits (1561), Expect = e-174
Identities = 307/307 (100%), Positives = 307/307 (100%)
Frame = +3
Query: 1 MATSLASQLSVGLRRPSPKLDSLNSQSTHSLFHSNLRLPISSSKPSRSVIAMAGSGKFFV 60
MATSLASQLSVGLRRPSPKLDSLNSQSTHSLFHSNLRLPISSSKPSRSVIAMAGSGKFFV
Sbjct: 93 MATSLASQLSVGLRRPSPKLDSLNSQSTHSLFHSNLRLPISSSKPSRSVIAMAGSGKFFV 272
Query: 61 GGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWV 120
GGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWV
Sbjct: 273 GGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWV 452
Query: 121 SKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIG 180
SKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIG
Sbjct: 453 SKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIG 632
Query: 181 ELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAI 240
ELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAI
Sbjct: 633 ELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAI 812
Query: 241 RDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSV 300
RDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSV
Sbjct: 813 RDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSV 992
Query: 301 TSKKVAA 307
TSKKVAA
Sbjct: 993 TSKKVAA 1013
>TC14860 similar to UP|TPIS_COPJA (P21820) Triosephosphate isomerase,
cytosolic (TIM) , partial (68%)
Length = 917
Score = 203 bits (517), Expect = 3e-53
Identities = 98/170 (57%), Positives = 123/170 (71%), Gaps = 2/170 (1%)
Frame = +3
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL--EPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K+V LN AK+ E V+VVV+PPFV++ VK + V+
Sbjct: 408 KFFVGGNWKCNGTTEQVKKIVGTLNEAKVPGEDIVEVVVSPPFVFLSFVKALLRSDFHVA 587
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGA+TGE+S E L + + WVILGHSERR ++ E EF+ K AYAL++ L
Sbjct: 588 AQNCWVHKGGAYTGEVSAEMLVNLDIPWVILGHSERRALLKESSEFVADKVAYALSQNLK 767
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGT 224
VIACIGE LE+REAG T V +Q KA A + +WDN+V+AYEPVWAIGT
Sbjct: 768 VIACIGETLEQREAGTTLAVVSEQTKAIAAKISNWDNVVLAYEPVWAIGT 917
>TC8012 similar to UP|TPIC_FRAAN (Q9M4S8) Triosephosphate isomerase,
chloroplast precursor (TIM) , partial (21%)
Length = 482
Score = 118 bits (295), Expect = 1e-27
Identities = 60/66 (90%), Positives = 61/66 (91%)
Frame = +3
Query: 242 DWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVT 301
DWL KNVSAEVASKTRIIYGG VNGG SELAKKEDIDGFLVGGASLKGPEFATIINSVT
Sbjct: 3 DWLPKNVSAEVASKTRIIYGGCVNGGTRSELAKKEDIDGFLVGGASLKGPEFATIINSVT 182
Query: 302 SKKVAA 307
S +VAA
Sbjct: 183 SPQVAA 200
>TC16258 similar to UP|TPIS_COPJA (P21820) Triosephosphate isomerase,
cytosolic (TIM) , partial (31%)
Length = 622
Score = 110 bits (275), Expect = 3e-25
Identities = 57/79 (72%), Positives = 63/79 (79%)
Frame = -3
Query: 225 GKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVG 284
GKVA+P QAQEVHA +R W+ NVSAEVAS RIIYGGSVNG NS ELA + D+DGFLVG
Sbjct: 620 GKVATPAQAQEVHADLRKWIHDNVSAEVASTVRIIYGGSVNGANSKELAGQADVDGFLVG 441
Query: 285 GASLKGPEFATIINSVTSK 303
GASLK PEF IIN+ T K
Sbjct: 440 GASLK-PEFIDIINAATVK 387
>TC8256 homologue to UP|AAR12195 (AAR12195) Molecular chaperone Hsp90-1,
partial (47%)
Length = 1251
Score = 32.3 bits (72), Expect = 0.10
Identities = 23/95 (24%), Positives = 43/95 (45%)
Frame = +2
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K D ++ ++ + F +E+LK G + + + + + IG+
Sbjct: 245 YVTRMKEGQNDIYYITGESKKAVENSPF-----LEKLKKKGYEVLYMVDAIDEYAIGQLK 409
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + FD
Sbjct: 410 EFEGKKLVSATKEGLKLEESEDEKKKQEELKEKFD 514
>TC15609 homologue to UP|HS82_ARATH (P55737) Heat shock protein 81-2
(HSP81-2), partial (42%)
Length = 1117
Score = 31.2 bits (69), Expect = 0.23
Identities = 22/95 (23%), Positives = 44/95 (46%)
Frame = +3
Query: 99 YIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKD 158
Y+ ++K +D ++ ++ + F +E+LK G + + + + + +G+
Sbjct: 138 YVTRMKEGQSDIYYITGESKKSVENSPF-----LEKLKKKGYEVLYMVDAIDEYAVGQLK 302
Query: 159 EFIGKKAAYALTEGLGVIACIGELLEEREAGKTFD 193
EF GKK A EGL + E ++ E + FD
Sbjct: 303 EFEGKKLVSATKEGLKLDETEDEKKKKEELKEKFD 407
>BP085060
Length = 376
Score = 28.1 bits (61), Expect = 2.0
Identities = 14/37 (37%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Frame = -2
Query: 33 HSNLRLPISSSKPSRSVIAMAGSGK-FFVGGNWKCNG 68
H+N+ I + KPSR + S + FFV +W C G
Sbjct: 312 HANMVXDIEALKPSRLPLTWWSSNRIFFVTSSW*CKG 202
>TC13617 similar to UP|Q9FKY1 (Q9FKY1) Similarity to ribonucleoprotein F,
partial (35%)
Length = 474
Score = 27.7 bits (60), Expect = 2.5
Identities = 13/47 (27%), Positives = 23/47 (48%)
Frame = +2
Query: 40 ISSSKPSRSVIAMAGSGKFFVGGNWKCNGTKDSISKLVADLNSAKLE 86
+S + R + G G+ +V WKC+G + I ++ + SA E
Sbjct: 167 LSFQEQCRLSLLCRGIGRTWVADTWKCSGVRSRIITMLLLVRSATKE 307
>TC14207 similar to UP|RK15_PEA (P31165) 50S ribosomal protein L15,
chloroplast precursor (CL15) (Fragment), partial (89%)
Length = 1201
Score = 27.3 bits (59), Expect = 3.3
Identities = 13/36 (36%), Positives = 16/36 (44%)
Frame = -1
Query: 33 HSNLRLPISSSKPSRSVIAMAGSGKFFVGGNWKCNG 68
H N R P + KP + K+F G KCNG
Sbjct: 1171 HYNRRFPANIGKPKFTFYQALNFSKYFGDG*TKCNG 1064
>TC12798 similar to UP|O39876 (O39876) HBc antigen, partial (8%)
Length = 735
Score = 26.9 bits (58), Expect = 4.3
Identities = 15/47 (31%), Positives = 23/47 (48%)
Frame = +2
Query: 225 GKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSE 271
G A EQ +E A++R W + S + A++ GGSV+ E
Sbjct: 17 GWSAPLEQPREDAASVRTWKSSSGSVKAAARIAATSGGSVSARKKVE 157
>TC15236 homologue to UP|GCST_PEA (P49364) Aminomethyltransferase,
mitochondrial precursor (Glycine cleavage system T
protein) (GCVT) , complete
Length = 1457
Score = 26.6 bits (57), Expect = 5.7
Identities = 20/45 (44%), Positives = 23/45 (50%), Gaps = 3/45 (6%)
Frame = +1
Query: 186 REAGKTFDV---CFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKV 227
RE G FDV C LK DAVP + +VIA A GTG +
Sbjct: 304 RENGSLFDVSHMCGLSLKG-KDAVPFLEKLVIADVASLAPGTGSL 435
>TC8417 similar to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimerase
(Galactowaldenase) (UDP-galactose 4-epimerase) ,
partial (65%)
Length = 789
Score = 25.8 bits (55), Expect = 9.7
Identities = 11/34 (32%), Positives = 18/34 (52%)
Frame = +3
Query: 28 THSLFHSNLRLPISSSKPSRSVIAMAGSGKFFVG 61
TH + N +SSS P + ++ G+G F+G
Sbjct: 48 THQILSPNSNKMVSSSSPQQKILVTGGAG--FIG 143
>TC18994
Length = 514
Score = 25.8 bits (55), Expect = 9.7
Identities = 25/100 (25%), Positives = 46/100 (46%), Gaps = 6/100 (6%)
Frame = +1
Query: 71 DSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWVSKGGAFTGEI 130
DS + V + +++KLE + D +VA +Q+K +T +E S + + V A E
Sbjct: 124 DSFANGVEEESTSKLERERDELVAENVAKKEQIK-KLTAELEESRKMTDVIAARASDLET 300
Query: 131 SVEQLKDHGVKWVILGHSERRHI------IGEKDEFIGKK 164
+ +L+ + G R + +G+K+ IG K
Sbjct: 301 ELSRLQHDMGSEMSAGEDARAKVADLTKAVGDKESRIGVK 420
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,876,949
Number of Sequences: 28460
Number of extensions: 62008
Number of successful extensions: 274
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 272
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 273
length of query: 307
length of database: 4,897,600
effective HSP length: 90
effective length of query: 217
effective length of database: 2,336,200
effective search space: 506955400
effective search space used: 506955400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0012b.1


