

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012a.7
(1015 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
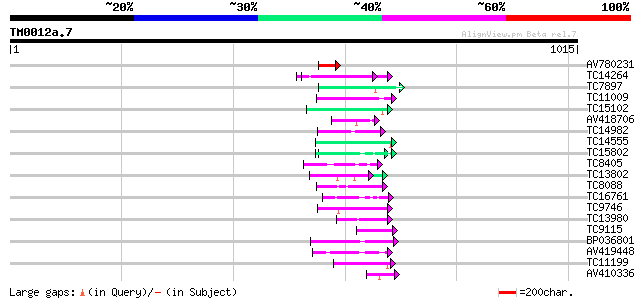
Score E
Sequences producing significant alignments: (bits) Value
AV780231 66 3e-11
TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), par... 52 5e-07
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 51 8e-07
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 50 1e-06
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 48 9e-06
AV418706 47 2e-05
TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%) 46 3e-05
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 45 6e-05
TC15802 similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber... 45 8e-05
TC8405 weakly similar to UP|O04083 (O04083) Lysophospholipase is... 44 1e-04
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 44 2e-04
TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like arabinogalac... 43 2e-04
TC16761 similar to GB|AAL32703.1|17065098|AY062625 nucleotide su... 43 2e-04
TC9746 43 2e-04
TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.3... 43 3e-04
TC9115 similar to PIR|T00425|T00425 photolyase/blue-light recept... 43 3e-04
BP036801 42 4e-04
AV419448 42 4e-04
TC11199 42 5e-04
AV410336 42 6e-04
>AV780231
Length = 530
Score = 65.9 bits (159), Expect = 3e-11
Identities = 32/39 (82%), Positives = 34/39 (87%)
Frame = -3
Query: 553 SNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPAS 591
SNPGAQPTTAAA KGKNV EAS+AAA EPT VPASTP +
Sbjct: 477 SNPGAQPTTAAARKGKNVNEASVAAAIEPTTVPASTPVT 361
Score = 37.7 bits (86), Expect = 0.009
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = -3
Query: 407 AKSSFARKYFRVAVHPA 423
AKSSFARKYFRVAVHPA
Sbjct: 528 AKSSFARKYFRVAVHPA 478
>TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), partial (76%)
Length = 1277
Score = 52.0 bits (123), Expect = 5e-07
Identities = 36/136 (26%), Positives = 57/136 (41%)
Frame = +2
Query: 523 KKRGGAENQAESTKAPKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPT 582
K + A+ A +TKA K + ++ + A + A+P AA PK K A+A AA +
Sbjct: 530 KPKAKAKPAAAATKA-KAKPAPKSKAAATKTTAKAKPAAAAKPKAKPAAKAKPAAKAKAV 706
Query: 583 AVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQ 642
A PA AS A A + + V K AA + + + ++ P +
Sbjct: 707 AAPAKAKASPAKPKAKAKSKTAPRMNVNPPVPKAKPAAKAKPAARPAKASRTSTRTTPGK 886
Query: 643 EAPPSPPPTRDAGSMP 658
+ PP P P + P
Sbjct: 887 KVPPPPKPAPKKAATP 934
Score = 45.4 bits (106), Expect = 4e-05
Identities = 48/177 (27%), Positives = 72/177 (40%), Gaps = 4/177 (2%)
Frame = +2
Query: 513 SPKFNTEGDAKKRGGAENQAESTKA---PKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKN 569
S K + A K A+ + + A PK + + ++ A + P + AA K
Sbjct: 443 SAKSSAAAAASKPAAAKKKPQPAAAKSKPKPKAKAKPAAAATKAKAKPAPKSKAAAT-KT 619
Query: 570 VAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEK 629
A+A AAA +P A PA+ A AVAA + A+ A KA A S T
Sbjct: 620 TAKAKPAAAAKPKAKPAAKAKPAAKAKAVAAPAKAKASPAK-----PKAKAKSKT-APRM 781
Query: 630 EKENETPKSPPHQEAPPSPPPTR-DAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPA 685
PK+ P +A P+ P + S + P + P A A +++APA
Sbjct: 782 NVNPPVPKAKPAAKAKPAARPAKASRTSTRTTPGKKVPPPPKPAPKKAATPVKKAPA 952
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 51.2 bits (121), Expect = 8e-07
Identities = 45/163 (27%), Positives = 65/163 (39%), Gaps = 8/163 (4%)
Frame = +3
Query: 553 SNPGAQPTTAAAPKGKNVAEA---SIAAATEPTAVP-ASTPASAGATAAVAAESSVGATA 608
S P P TA P + S A A P P A TP SA + + + S+ A +
Sbjct: 579 SPPSGSPPTAGEPSPAGTPPSVLPSPAGAPVPAGGPSAGTPPSASTSPSPSPVSAPPAAS 758
Query: 609 ASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTR----DAGSMPSPPHQG 664
+ G + ++ +S++P G + +P PP+P PT + S P G
Sbjct: 759 PAPGAESPSSSPTSNSPAGGPTATSPSPAGDSPAGGPPAPSPTTGDTPPSASGPDVSPSG 938
Query: 665 EKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFL 707
S A T S ++ AP P G SS + PS FL
Sbjct: 939 TPSSLPADTPSSSSNNSTAPPPGNGGSS--------VVPSSFL 1043
Score = 31.6 bits (70), Expect = 0.66
Identities = 22/62 (35%), Positives = 24/62 (38%), Gaps = 7/62 (11%)
Frame = +3
Query: 638 SPPHQEAPPSPPPTRDAGSMP-------SPPHQGEKSCPGAATTSEAAQIEQAPAPEVGS 690
SP + PPSPPP P SPP GE S P S AP P G
Sbjct: 510 SPRGTQPPPSPPPKSSPSPSPKASPPSGSPPTAGEPS-PAGTPPSVLPSPAGAPVPAGGP 686
Query: 691 SS 692
S+
Sbjct: 687 SA 692
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 50.4 bits (119), Expect = 1e-06
Identities = 37/147 (25%), Positives = 70/147 (47%), Gaps = 4/147 (2%)
Frame = -2
Query: 549 GAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASA-GATAAVAAESSVGAT 607
G +S+P + +++ P ++ A P+++ S+P+S+ G+ + A+ SS AT
Sbjct: 441 GLSSSSPPSSESSSPEPSLSDLFLFDGTPAFSPSSLSLSSPSSSSGSKSLDASSSSAAAT 262
Query: 608 AASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPP---TRDAGSMPSPPHQG 664
+ K+++SS + + + SPP PPS PP + + S SPP
Sbjct: 261 VRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPR-PPSTPPSSSSSSSSSSSSPP--- 94
Query: 665 EKSCPGAATTSEAAQIEQAPAPEVGSS 691
PGA+++S ++ +P+ SS
Sbjct: 93 ----PGASSSSSSSPSSSSPSSSSSSS 25
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 47.8 bits (112), Expect = 9e-06
Identities = 37/160 (23%), Positives = 63/160 (39%), Gaps = 7/160 (4%)
Frame = +1
Query: 532 AESTKAPKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPAS 591
A +T K + T+ P ++PT AP ++S ++ P+ ++TP S
Sbjct: 181 ATTTSTTKAATNSSPPESRSTTPPSSEPTCLLAPASPPTPKSSHSSTPPPSRSSSTTPKS 360
Query: 592 AGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEK-ENETPKSPPHQEAPPSPPP 650
AT+ A A+ A+ +K + T + ++ P + P + P P
Sbjct: 361 KNATSLTAPSCPPPASPAATAPAPSKTPPNPPTNSSKPSSCKSSLPNTTPSSLSSAPPSP 540
Query: 651 TRDAGSMPSPPHQGEK------SCPGAATTSEAAQIEQAP 684
T S PS P EK S P +T+S ++ P
Sbjct: 541 TTSPISTPSSPENQEKPASTPPSAPPRSTSSSSSSPINTP 660
Score = 40.0 bits (92), Expect = 0.002
Identities = 49/181 (27%), Positives = 69/181 (38%), Gaps = 11/181 (6%)
Frame = +1
Query: 530 NQAESTKAPKRRRLTRTS-SGAGTSNPGAQPTTAAAPKGKNVA-EASIAAATEPTAVPAS 587
+Q E TK R R+S + P TAAA N + EA+ + ++EP +
Sbjct: 10 SQFEKTKQTNRYSSVRSSHQQQWQQHLQTTPKTAAATFL*NPSLEATASPSSEPCTTATT 189
Query: 588 TPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGE-------KEKENETPKSPP 640
T + AT + ES T S+ A AS TP + TPKS
Sbjct: 190 TSTTKAATNSSPPESR-STTPPSSEPTCLLAPASPPTPKSSHSSTPPPSRSSSTTPKSKN 366
Query: 641 HQE--APPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLP 698
AP PPP A + P+P K+ P T S ++ P SS + P
Sbjct: 367 ATSLTAPSCPPPASPAATAPAP----SKTPPNPPTNSSKPSSCKSSLPNTTPSSLSSAPP 534
Query: 699 N 699
+
Sbjct: 535 S 537
Score = 38.5 bits (88), Expect = 0.005
Identities = 41/160 (25%), Positives = 60/160 (36%), Gaps = 6/160 (3%)
Frame = +1
Query: 514 PKFNTEGDAKKRGGAENQAESTKAPKRRRLTRTSSGAGTSNPGAQPTTAAAPKG-----K 568
P ++ K + A S P T + NP PT ++ P
Sbjct: 328 PSRSSSTTPKSKNATSLTAPSCPPPASPAATAPAPSKTPPNP---PTNSSKPSSCKSSLP 498
Query: 569 NVAEASIAAAT-EPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIG 627
N +S+++A PT P STP+S + +T SA +T +++SS PI
Sbjct: 499 NTTPSSLSSAPPSPTTSPISTPSSP------ENQEKPASTPPSAPPRSTSSSSSS--PIN 654
Query: 628 EKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKS 667
K + K AP S PP + SPP KS
Sbjct: 655 TPRKPSSATK------APASSPPGSELSLHHSPPSASRKS 756
>AV418706
Length = 374
Score = 47.0 bits (110), Expect = 2e-05
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 4/89 (4%)
Frame = +1
Query: 577 AATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKA----AASSDTPIGEKEKE 632
A T P VP P +A A AA SS G++++S+ +++ A +++S T +
Sbjct: 94 APTAPPTVPVLHPLAAAAATVAAAHSSAGSSSSSSSSSSSSAPPAPSSTSSTTLNALPSP 273
Query: 633 NETPKSPPHQEAPPSPPPTRDAGSMPSPP 661
+ PP PP PPP+ + + SPP
Sbjct: 274 SRHSNFPPSNSPPP-PPPSPPSSNSHSPP 357
>TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%)
Length = 1036
Score = 46.2 bits (108), Expect = 3e-05
Identities = 32/123 (26%), Positives = 53/123 (43%), Gaps = 1/123 (0%)
Frame = +3
Query: 552 TSNPGAQPTTAAAPKGKNVAEASIAAATEPTAV-PASTPASAGATAAVAAESSVGATAAS 610
T P P A P ++ +T PTA+ A P++A AA +A S+ G +++S
Sbjct: 45 TLPPKPPPPQTAPPPPTQLSPPQRPNSTAPTALLTALNPSTAAPAAAASAPSASG*SSSS 224
Query: 611 AGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPG 670
+ +++SS + + P +PP P + P + PSPP+ S P
Sbjct: 225 S------SSSSSSAVLASPSTSSTAPTTPPSPSPPSNSPTSTSPPPQPSPPNSTSPSPPQ 386
Query: 671 AAT 673
T
Sbjct: 387 TPT 395
Score = 28.5 bits (62), Expect = 5.6
Identities = 19/49 (38%), Positives = 22/49 (44%), Gaps = 7/49 (14%)
Frame = +3
Query: 645 PPSPPPTRDAGSMP---SPPHQGEKSCPGAATT----SEAAQIEQAPAP 686
PP PPP + A P SPP + + P A T S AA A AP
Sbjct: 51 PPKPPPPQTAPPPPTQLSPPQRPNSTAPTALLTALNPSTAAPAAAASAP 197
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 45.1 bits (105), Expect = 6e-05
Identities = 38/155 (24%), Positives = 61/155 (38%), Gaps = 10/155 (6%)
Frame = +2
Query: 548 SGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAG---ATAAVAAESSV 604
+ G +P PTT+ AP + A ++ P A + P + A+ A+SS
Sbjct: 170 ASVGAQSPSNSPTTSPAPPTPTTPQPPAAQSSPPPAQSSPPPVQSSPPPASTPPPAQSSP 349
Query: 605 GATAASAGVNATKAAASSDTPIGEKEKE-NETPKSPPHQEAPPSPPPTRDAGSMPSP-PH 662
++ V +T A + TP + P +PP A P P T + P+P P
Sbjct: 350 PPVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAATPPPALTPVPATSPAPAPA 529
Query: 663 QGEKSCPGAATTSEAAQI-----EQAPAPEVGSSS 692
+ + P A A + +AP P + S S
Sbjct: 530 KVKSKSPAPAPAPALAPVVSLSPSEAPGPSLSSLS 634
>TC15802 similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber hypocotyls
(Arabinogalactan protein), partial (26%)
Length = 795
Score = 44.7 bits (104), Expect = 8e-05
Identities = 39/145 (26%), Positives = 58/145 (39%), Gaps = 1/145 (0%)
Frame = +2
Query: 548 SGAGTSNPGAQPTTA-AAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGA 606
+G G +P A PTT+ A P +A P A+TPA++ +TA A ++
Sbjct: 92 AGVGGQSPTAAPTTSPATPSATTPVSTPVAPVASPKPA-ATTPAASPSTATAPAPATTTP 268
Query: 607 TAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEK 666
+S P P S P ++PP+P PT A + +P E
Sbjct: 269 VTPVTSPPPAAVPVASPPPAA-------VPVSSPPAKSPPAPAPT--AVPVAAPVTTPEV 421
Query: 667 SCPGAATTSEAAQIEQAPAPEVGSS 691
P + T + A AP+P V S
Sbjct: 422 PAPAPSKTKKDA--APAPSPVVPDS 490
Score = 44.3 bits (103), Expect = 1e-04
Identities = 33/130 (25%), Positives = 51/130 (38%), Gaps = 2/130 (1%)
Frame = +2
Query: 553 SNPGAQPTTAAAPKGKNVAEASIAAATEPTAVP-ASTPASAGATAAVAAESSVGATAASA 611
+ P A P+TA AP + + P AVP AS P +A ++ A+S +
Sbjct: 209 TTPAASPSTATAPAPATTTPVTPVTSPPPAAVPVASPPPAAVPVSSPPAKSPPAPAPTAV 388
Query: 612 GVNATKAAASSDTPIGEKEKENETP-KSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPG 670
V A P K K++ P SP ++PP+ P P P G +
Sbjct: 389 PVAAPVTTPEVPAPAPSKTKKDAAPAPSPVVPDSPPAGAPGPSDAVSPGPDGAGTANDES 568
Query: 671 AATTSEAAQI 680
A T + ++
Sbjct: 569 GAETMRSLKM 598
>TC8405 weakly similar to UP|O04083 (O04083) Lysophospholipase isolog,
25331-24357 (At1g11090), partial (18%)
Length = 552
Score = 43.9 bits (102), Expect = 1e-04
Identities = 37/142 (26%), Positives = 58/142 (40%), Gaps = 1/142 (0%)
Frame = +3
Query: 527 GAENQAESTKAPKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPA 586
GA + +T PK +S T+ P++ + PK K P++ P
Sbjct: 75 GATCRKRNTTPPKESATPNPTSTLPTARSSPSPSSPSTPKSK-----------PPSS*PT 221
Query: 587 ST-PASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAP 645
+T P AG++ A+ S GAT +S ++ AA P + SPP P
Sbjct: 222 ATAPTPAGSSRRSASTSPPGATPSSPPTSSATAA-----PTASAATSATSTASPP----P 374
Query: 646 PSPPPTRDAGSMPSPPHQGEKS 667
SP + A + P+PP + S
Sbjct: 375 RSPSSSTSAAASPTPPFRPSSS 440
Score = 33.1 bits (74), Expect = 0.23
Identities = 28/110 (25%), Positives = 44/110 (39%), Gaps = 6/110 (5%)
Frame = +3
Query: 589 PASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAP-PS 647
P ++GAT + +A + A SS +P +++ P S P AP P+
Sbjct: 63 PLTSGATCRKRNTTPPKESATPNPTSTLPTARSSPSPSSPSTPKSKPPSS*PTATAPTPA 242
Query: 648 PPPTRDA-----GSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 692
R A G+ PS P + A+ + +A +P P SSS
Sbjct: 243 GSSRRSASTSPPGATPSSPPTSSATAAPTASAATSATSTASPPPRSPSSS 392
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/142 (24%), Positives = 54/142 (37%), Gaps = 4/142 (2%)
Frame = +2
Query: 538 PKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAA 597
P LT+ S +SNP +T ++P + A + P+S+PASA ++
Sbjct: 59 PLHSTLTKPPSQI-SSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPASS 235
Query: 598 VAAESSVGATAASAGVNAT----KAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRD 653
+ +ASA ++T A S +P + +P P A +
Sbjct: 236 PTKSPKPSSPSASAPPSSTPLTPSTATSESSPTATSSSSSASPAPPTSFSALSRALELKA 415
Query: 654 AGSMPSPPHQGEKSCPGAATTS 675
S PSPP S P A S
Sbjct: 416 LASSPSPPSSPPPSPPSAICMS 481
Score = 43.5 bits (101), Expect = 2e-04
Identities = 30/120 (25%), Positives = 50/120 (41%), Gaps = 6/120 (5%)
Frame = +2
Query: 538 PKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVP------ASTPAS 591
P + T +S + T P P ++ + + + AS + A+ PT P AS P S
Sbjct: 107 PNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPASSPTKSPKPSSPSASAPPS 286
Query: 592 AGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPT 651
+ A S TA S+ +A+ A +S + + + SP +PP PP+
Sbjct: 287 STPLTPSTATSESSPTATSSSSSASPAPPTSFSALSRALELKALASSPSPPSSPPPSPPS 466
Score = 35.4 bits (80), Expect = 0.046
Identities = 46/172 (26%), Positives = 66/172 (37%), Gaps = 2/172 (1%)
Frame = +2
Query: 79 LHNRPIVRSNSPLHATHLPKTIMTSNLPH--ASNTRHPSSFSPSPIKTLLPHCHPFPRPL 136
LH + + N PLH+T S+ P+ S T P+S +P P P P P
Sbjct: 26 LHFQSTILPNKPLHSTLTKPPSQISSNPNKTTSTTSSPTSTTPKP--------SPSPAPS 181
Query: 137 FTSLIAYQTPSVHFRSGAVAYHPFRYPLFTSYSGESHCPYLYIAHILIFSFLSLHCLLKM 196
T + PS S + A P + P +S S + + S
Sbjct: 182 ST----LRAPSSSPASASPASSPTKSPKPSSPSASAPPSSTPLTPSTATS---------- 319
Query: 197 ASNPTSPNHSSSSSESDPDSTAAGGLRLEVPEIPAHRLKTYATLPLPVQVAP 248
S+PT+ + SSS+S + P S +A LE LK A+ P P P
Sbjct: 320 ESSPTATSSSSSASPAPPTSFSALSRALE--------LKALASSPSPPSSPP 451
>TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (69%)
Length = 1527
Score = 43.1 bits (100), Expect = 2e-04
Identities = 35/129 (27%), Positives = 53/129 (40%), Gaps = 3/129 (2%)
Frame = +2
Query: 550 AGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAA 609
A T+ P P T +P + + + A TA P+ P+S T S+ +T+
Sbjct: 140 AATTLPAFSPNTPTSPPSTTTSPSPTSPARSTTAKPS--PSSPSTTPPCPPSST--STSP 307
Query: 610 SAGVNATKAAASSDTPIGEKEKEN---ETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEK 666
S + + SS T + K E P SPP PP+PPP A S + +
Sbjct: 308 SPP*KTSSPSTSSSTTLAPKSSTR*PTEPPSSPPCSR-PPAPPPEPPATSTSPTSKEAK* 484
Query: 667 SCPGAATTS 675
+ P TT+
Sbjct: 485 ASPLRTTTA 511
>TC16761 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;}, partial
(40%)
Length = 794
Score = 43.1 bits (100), Expect = 2e-04
Identities = 33/127 (25%), Positives = 56/127 (43%), Gaps = 1/127 (0%)
Frame = +1
Query: 561 TAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAA 620
T A P NV A+ +T + ++ P ++ + ++ SS +++ S + +
Sbjct: 187 TQAKP*SWNVTTATSVKSTAQSF--STLPPNSSSAPPSSSHSSSSSSSPSTTLPSPPTPP 360
Query: 621 SSDTPIGEKEKENETP-KSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQ 679
++ TP TP SPPH A P P R + P PPH +CP ++ A+
Sbjct: 361 TTSTP---------TPTSSPPHSAAEP---PGRSKSATP-PPHAAPTACPSSSPAQSASS 501
Query: 680 IEQAPAP 686
AP+P
Sbjct: 502 AHTAPSP 522
>TC9746
Length = 637
Score = 43.1 bits (100), Expect = 2e-04
Identities = 34/141 (24%), Positives = 62/141 (43%), Gaps = 7/141 (4%)
Frame = +2
Query: 552 TSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPAS-------TPASAGATAAVAAESSV 604
T +P ++P P + A +S A+ +EP + P S P S ++ ++ + +
Sbjct: 170 TPSPTSKPKCQPPPHPSSTATSSPASNSEPPSGPNSPARWGT*APTSPSSSPSL*STTLT 349
Query: 605 GATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQG 664
+S+ + T ASS + + +P SP + P SP P + PSPP+
Sbjct: 350 SPPHSSSPHSTTSPLASSSASPCQSSP*SPSPPSPSPKH-PHSPSPRSPPPASPSPPYFS 526
Query: 665 EKSCPGAATTSEAAQIEQAPA 685
+ P + +S A + Q+ A
Sbjct: 527 SSAPPVSCPSSTATSLCQSSA 589
>TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.33), partial
(12%)
Length = 565
Score = 42.7 bits (99), Expect = 3e-04
Identities = 29/102 (28%), Positives = 43/102 (41%), Gaps = 2/102 (1%)
Frame = +2
Query: 585 PASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEA 644
PAST SA +++ + +++AS + SS +P G P PP Q+
Sbjct: 242 PASTTPSATRASSLGIPNPPTSSSASKTPSPCSPPTSS-SPTGSSSHSTSNPPHPPPQQQ 418
Query: 645 PPSPPPTRDAGSMPSPP--HQGEKSCPGAATTSEAAQIEQAP 684
P SPPP P+ P H + P A S A++ P
Sbjct: 419 PFSPPPPPPPPPSPTAPSSHPRLPAAPAAGRISSASKSSTTP 544
>TC9115 similar to PIR|T00425|T00425 photolyase/blue-light receptor (PHR2)
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (55%)
Length = 1018
Score = 42.7 bits (99), Expect = 3e-04
Identities = 22/74 (29%), Positives = 31/74 (41%)
Frame = +1
Query: 621 SSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQI 680
S P + P SPP AP +P ++P+PPH G+ATTS +A
Sbjct: 292 SPPPPHPRRPNPPSNPPSPPTPSAPLTP*APTAPSTLPTPPHSAAPPSSGSATTSASATT 471
Query: 681 EQAPAPEVGSSSYY 694
+ P S +Y
Sbjct: 472 RLSTPPTTNRSQFY 513
Score = 30.0 bits (66), Expect = 1.9
Identities = 21/70 (30%), Positives = 25/70 (35%), Gaps = 12/70 (17%)
Frame = -2
Query: 625 PIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSP-PHQGEKSC-----------PGAA 672
P EK K P +PP PP+ S P P PH + C P
Sbjct: 861 PPTEKANPQHGTKCSPKSTSPPHSPPSSPPQSSPPPSPHHAKPLCEHTQRQHQQPSPAPP 682
Query: 673 TTSEAAQIEQ 682
T S A Q+ Q
Sbjct: 681 TQSPACQL*Q 652
>BP036801
Length = 540
Score = 42.4 bits (98), Expect = 4e-04
Identities = 45/158 (28%), Positives = 65/158 (40%), Gaps = 1/158 (0%)
Frame = +2
Query: 539 KRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAV 598
K + L +SS S A P AAA G + + A+ T +T + +TA
Sbjct: 8 KNKPLLLSSSP*SPSCCSASPQHAAAQGGTGFSYNAA*ASQPCTCRS*TTTRWSSSTAPT 187
Query: 599 AAE-SSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSM 657
+A +S TAA+A T ++S+ P + SPP A PT A
Sbjct: 188 SAHPTSPSPTAAAASTPVTWPSSSTAPPTPS------STTSPPTLSALSPSKPTHGAPPA 349
Query: 658 PSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYN 695
PSPP S P A+TT+ + PA S++ N
Sbjct: 350 PSPP-TAPSSKPAASTTATTSSAPSPPAHNTTSATGSN 460
>AV419448
Length = 382
Score = 42.4 bits (98), Expect = 4e-04
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 3/147 (2%)
Frame = +2
Query: 542 RLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPAS-TPASAGATAAVAA 600
RL+ + + +S+P P+ + P + T P+ +S T ASA ++ A
Sbjct: 8 RLSTSPAAPSSSHPPPSPSQPSLPS------PPLHPPTPPSPTASSWTSASAPPPSSPTA 169
Query: 601 ESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPS--PPPTRDAGSMP 658
S SA ++ A SS +P +P S P PP+ PPPT+ S
Sbjct: 170 TPSAPTPPLSAALSLASTATSSPSP---------SPISNPSASLPPTPPPPPTKTPSSTS 322
Query: 659 SPPHQGEKSCPGAATTSEAAQIEQAPA 685
S P AAT+S AA+++ A
Sbjct: 323 SSP---------AATSSPAARVDPTRA 376
Score = 29.3 bits (64), Expect = 3.3
Identities = 25/99 (25%), Positives = 37/99 (37%), Gaps = 10/99 (10%)
Frame = +2
Query: 615 ATKAAASSDTPIGEKEKENETPKSPPHQEAPPSP----------PPTRDAGSMPSPPHQG 664
+T AA S + + P P H PPSP PP + PS P
Sbjct: 14 STSPAAPSSSHPPPSPSQPSLPSPPLHPPTPPSPTASSWTSASAPPPSSPTATPSAP--- 184
Query: 665 EKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEP 703
+ P +A S A+ +P+P S+ ++ P P
Sbjct: 185 --TPPLSAALSLASTATSSPSPSPISNPSASLPPTPPPP 295
>TC11199
Length = 743
Score = 42.0 bits (97), Expect = 5e-04
Identities = 34/118 (28%), Positives = 49/118 (40%), Gaps = 7/118 (5%)
Frame = +1
Query: 580 EPTAVPASTPASAGATAAVAAESSVGATAASA-GVNATKAAASSDTPIGEKEKENETPKS 638
+P++ S+PAS + +A S + S T +SS + I + P S
Sbjct: 157 DPSSARPSSPASQSHSTTTSATSLAPKSPRSKRSARRTTRTSSSPSTISDPSSPTSNPSS 336
Query: 639 PPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATT----SEAAQI--EQAPAPEVGS 690
PP PS P+ S PS P PG +T+ SE+A + APAP S
Sbjct: 337 PPSLTPTPSSSPSLALSSPPSTPSSKRGMYPGTSTSPSTRSESAFS*WKIAPAPTATS 510
>AV410336
Length = 358
Score = 41.6 bits (96), Expect = 6e-04
Identities = 22/67 (32%), Positives = 30/67 (43%), Gaps = 7/67 (10%)
Frame = +1
Query: 639 PPHQEAPPSPPPTRDAGSMPSP----PHQGEKSCPGAATTSEAAQIEQAPAPE---VGSS 691
PPH PP+P P + S P+P PH G S P T + + P P SS
Sbjct: 64 PPHAPPPPTPNPKSPSSSSPTPTSPSPHSGRNSSPATTTFTTSTSTPTPPPPSSLPAASS 243
Query: 692 SYYNMLP 698
+ ++LP
Sbjct: 244 TAASLLP 264
Score = 33.5 bits (75), Expect = 0.17
Identities = 18/54 (33%), Positives = 26/54 (47%), Gaps = 2/54 (3%)
Frame = +1
Query: 641 HQEAP-PSPP-PTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 692
H+ +P PSPP PT S PPH P + S ++ +P+P G +S
Sbjct: 1 HKPSPHPSPPTPTTSTSSTAPPPHAPPPPTPNPKSPSSSSPTPTSPSPHSGRNS 162
Score = 27.7 bits (60), Expect = 9.6
Identities = 22/92 (23%), Positives = 35/92 (37%)
Frame = +1
Query: 534 STKAPKRRRLTRTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAG 593
ST P T + S+ PT+ + G+N + A+ T + P+S
Sbjct: 52 STAPPPHAPPPPTPNPKSPSSSSPTPTSPSPHSGRNSSPATTTFTTSTSTPTPPPPSSLP 231
Query: 594 ATAAVAAESSVGATAASAGVNATKAAASSDTP 625
A ++ AA A ++ AASS P
Sbjct: 232 AASSTAASLLPKKPTAPPQHSSPPPAASSPPP 327
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.129 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,256,073
Number of Sequences: 28460
Number of extensions: 297084
Number of successful extensions: 5234
Number of sequences better than 10.0: 434
Number of HSP's better than 10.0 without gapping: 3260
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4387
length of query: 1015
length of database: 4,897,600
effective HSP length: 99
effective length of query: 916
effective length of database: 2,080,060
effective search space: 1905334960
effective search space used: 1905334960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0012a.7


