

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.5
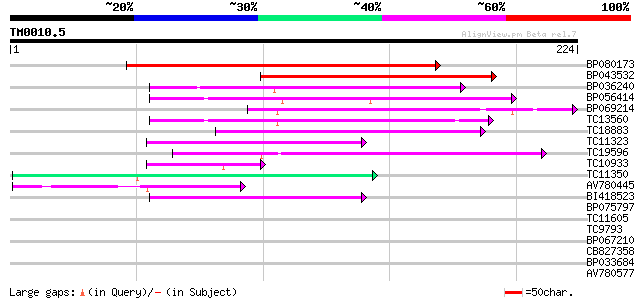
(224 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP080173 159 5e-40
BP043532 98 1e-21
BP036240 68 1e-12
BP056414 64 3e-11
BP069214 62 8e-11
TC13560 62 1e-10
TC18883 55 7e-09
TC11323 48 1e-06
TC19596 47 3e-06
TC10933 similar to UP|Q9LNS0 (Q9LNS0) F1L3.4, partial (6%) 47 3e-06
TC11350 similar to UP|Q9LJP7 (Q9LJP7) Genomic DNA, chromosome 3,... 45 8e-06
AV780445 43 5e-05
BI418523 42 9e-05
BP075797 38 0.001
TC11605 35 0.008
TC9793 35 0.010
BP067210 33 0.030
CB827358 33 0.052
BP033684 32 0.068
AV780577 32 0.12
>BP080173
Length = 382
Score = 159 bits (401), Expect = 5e-40
Identities = 75/124 (60%), Positives = 93/124 (74%)
Frame = -2
Query: 47 RAHGTGDLRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSIN 106
RA GT +RW P + WIKIN DGS++P SN AACGGV+ DH G ++ G S LG IN
Sbjct: 372 RASGTTFVRWPVPMEGWIKINIDGSFAPSSNCAACGGVMWDHEGTFITGASVRLGCYFIN 193
Query: 107 FAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFD 166
AELWGI+HGAK+A RGFKK+++ESDS+F V+LVN+GC PSKPA LVK+IK + F+
Sbjct: 192 HAELWGIFHGAKIAQARGFKKILVESDSSFVVSLVNQGCSPSKPATPLVKDIKSLFNCFE 13
Query: 167 QVHL 170
QVHL
Sbjct: 12 QVHL 1
>BP043532
Length = 465
Score = 97.8 bits (242), Expect = 1e-21
Identities = 46/93 (49%), Positives = 61/93 (65%)
Frame = -2
Query: 100 LGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIK 159
+G CSI AELWG+ HG + AL RGF K++IE+DSA + +NKGC P LV+
Sbjct: 464 VGVCSILHAELWGLVHGLRFALGRGFSKILIEADSAGTIEFLNKGCPVVHPCFPLVREFH 285
Query: 160 RTLSNFDQVHLKHVFREINSVADAFAKNGLGLL 192
+ +H KHV+RE+N VAD+FAKNGL +L
Sbjct: 284 HLVGQNCYIHWKHVYREVNFVADSFAKNGLRIL 186
>BP036240
Length = 567
Score = 67.8 bits (164), Expect = 1e-12
Identities = 39/128 (30%), Positives = 68/128 (52%), Gaps = 3/128 (2%)
Frame = -3
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRC---SINFAELWG 112
W P++ W K + DG+ S LA+CGGVLRD +++ GF +G ++ EL
Sbjct: 562 WTAPDEGWTKFDVDGALSR-DLLASCGGVLRDSRRHWIKGFCRNMGDLTYHNVFLVELSA 386
Query: 113 IYHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKH 172
I ++AL + V++ESD+ A++L++ + + P +L +I R + + +H
Sbjct: 385 IQSAVEIALSMDLQHVIVESDALEAIHLLSSENVHAHPFGSLAASILRMQRDHGSIVFQH 206
Query: 173 VFREINSV 180
RE NS+
Sbjct: 205 TPREANSL 182
>BP056414
Length = 606
Score = 63.5 bits (153), Expect = 3e-11
Identities = 46/149 (30%), Positives = 69/149 (45%), Gaps = 4/149 (2%)
Frame = -3
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSIN---FAELWG 112
W P + W K+N DG+ S A+CGGV+RD G++ GF+ LG + F EL
Sbjct: 562 WTRPPEGWFKVNVDGALSGDLQ-ASCGGVVRDAAGSWSKGFARSLGVLRWHXAFFVELMA 386
Query: 113 IYHGAKLALDRGFKKVMIESDSAFAVNLV-NKGCLPSKPAAALVKNIKRTLSNFDQVHLK 171
+ + +V+IESDS V L+ N + +I++ +S + +
Sbjct: 385 VQTAVDFIMSWDIPQVIIESDSQQVVELLQNFNASSPHQYGEIAWDIQQKISVHGSIVIT 206
Query: 172 HVFREINSVADAFAKNGLGLLMDFRIYDA 200
RE N +AD AK GL L +DA
Sbjct: 205 STPREANFLADYLAKVGLSLPFGTHSFDA 119
>BP069214
Length = 545
Score = 62.0 bits (149), Expect = 8e-11
Identities = 49/136 (36%), Positives = 68/136 (49%), Gaps = 6/136 (4%)
Frame = -2
Query: 95 GFSALLGRCS---INFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPA 151
G GR + I EL ++HG L DRGF++V+ +SDS A+ LV P
Sbjct: 445 GLQVFFGRGTL*EITNMELLAMWHGLDLVWDRGFRQVICQSDSTEALALVLGVPPPRHRY 266
Query: 152 AALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRI---YDAIPSFISVA 208
AAL+ +IK LS +V + H+ RE NS AD AK G + D + + +P +
Sbjct: 265 AALIWSIKDLLSREWEVSMTHMLREGNSCADFLAK--YGAMQDEPLVLPQEPLPGMTQL- 95
Query: 209 LMHDSCNTLYHRGGPP 224
L DS L+ RG PP
Sbjct: 94 LFWDSVEFLFERG*PP 47
>TC13560
Length = 571
Score = 61.6 bits (148), Expect = 1e-10
Identities = 42/139 (30%), Positives = 68/139 (48%), Gaps = 3/139 (2%)
Frame = +2
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCS---INFAELWG 112
W+ P+ W+KIN DG+ S A+C GVLR+ +G++L GF G S + EL
Sbjct: 146 WIKPDAGWVKINVDGAVSNCLQ-ASCRGVLRNADGSWLKGFRWNFGTFSSANVFLTELMA 322
Query: 113 IYHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKH 172
+ + A+ +V++ESDS V+ ++ L S + + +I + ++
Sbjct: 323 VKTAVEAAMSLDLARVIVESDSLEVVDFLHSTDLGSHRYSQIALDILHLQREHGALVFQY 502
Query: 173 VFREINSVADAFAKNGLGL 191
R NS+ D AK GL L
Sbjct: 503 ALRG-NSIVDYIAKFGLSL 556
>TC18883
Length = 821
Score = 55.5 bits (132), Expect = 7e-09
Identities = 32/107 (29%), Positives = 51/107 (46%)
Frame = +1
Query: 82 GGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLV 141
GG+ R+ + L FS +G AE+W I+ + ++R + + IESDS+ AV V
Sbjct: 16 GGIFRNSDNEILGFFSKHVGEGFAFEAEVWAIFEALTICVNRQLRNLTIESDSSLAVGWV 195
Query: 142 NKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNG 188
N + I ++ + + +KH+FRE N D AK G
Sbjct: 196 NNRSXRPWKLLNKLNQIDLWVAEVNCLSVKHIFREANGEVDKLAKRG 336
>TC11323
Length = 657
Score = 48.1 bits (113), Expect = 1e-06
Identities = 29/87 (33%), Positives = 40/87 (45%)
Frame = +2
Query: 55 RWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIY 114
RW+ P K+N DGS A GG+LRD + N++ GFS L I L I
Sbjct: 2 RWLKPVHGRFKLNFDGSSLGNRGNAGGGGLLRDGSSNFIFGFSIFLAVAQIMKLSLCAIL 181
Query: 115 HGAKLALDRGFKKVMIESDSAFAVNLV 141
G + G+ + IE DS V+ +
Sbjct: 182 EGLLVCKPLGYDGIGIECDSNIVVSWI 262
>TC19596
Length = 499
Score = 47.0 bits (110), Expect = 3e-06
Identities = 38/149 (25%), Positives = 67/149 (44%), Gaps = 1/149 (0%)
Frame = +1
Query: 65 KINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSA-LLGRCSINFAELWGIYHGAKLALDR 123
++++DG++ + GG++RD +G ++ GF A LG ++ AE+ + HG L +
Sbjct: 4 RMDSDGAFKHDDDRMGMGGIVRDAHGAWISGFYAGSLGGDALR-AEIAALKHGLTLLWNA 180
Query: 124 GFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADA 183
++ E D V + A+ + +I+ L V L +V RE N+ AD
Sbjct: 181 HVRRATCEVDCLDIVEALENDRYQFHALASELLDIRLLLDRDWTVTLAYVPREANAAADC 360
Query: 184 FAKNGLGLLMDFRIYDAIPSFISVALMHD 212
A G LL ++ P + L D
Sbjct: 361 LAGLGASLLCPLTCLESPPQELQPILARD 447
>TC10933 similar to UP|Q9LNS0 (Q9LNS0) F1L3.4, partial (6%)
Length = 596
Score = 46.6 bits (109), Expect = 3e-06
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 28/75 (37%)
Frame = +2
Query: 55 RWVPPNDDWIKINTDGSYSPVSNLAACGG----------------------------VLR 86
RW PP+ W+K+NTDGS++ S+L CGG +L+
Sbjct: 104 RWTPPSVGWVKLNTDGSFTRGSSLMGCGGLC*K**SDLFM*P*HNNFSF*DNWLFDSLLQ 283
Query: 87 DHNGNYLVGFSALLG 101
D +GN+ GFS+ G
Sbjct: 284 DRHGNWRAGFSSFEG 328
>TC11350 similar to UP|Q9LJP7 (Q9LJP7) Genomic DNA, chromosome 3, P1 clone:
MIG10, partial (18%)
Length = 1118
Score = 45.4 bits (106), Expect = 8e-06
Identities = 35/148 (23%), Positives = 59/148 (39%), Gaps = 4/148 (2%)
Frame = -3
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAH----GTGDLRWV 57
IW RN ++F+ P +I+ + QD T ++ + + D W
Sbjct: 969 IWKNRNRVIFQLEEPNPVNTIFQIRHMQQDANLMSTSQSNKQTTRERNINVTSARDRSWR 790
Query: 58 PPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGA 117
PP K+N+D S+ S G V+RD+ G + G ++ S +E + G
Sbjct: 789 PPPLGVYKLNSDASWKAPSPSCTVGVVIRDNLGLLISGTASRPLAPSPIVSEALALREGL 610
Query: 118 KLALDRGFKKVMIESDSAFAVNLVNKGC 145
LA G +++ ESD + C
Sbjct: 609 ILARSLGLDRILSESDCQPLIEACRSSC 526
>AV780445
Length = 504
Score = 42.7 bits (99), Expect = 5e-05
Identities = 30/98 (30%), Positives = 42/98 (42%), Gaps = 6/98 (6%)
Frame = +1
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGTGD------LR 55
+W RN +VFE +P V +KL + +T + DA GD R
Sbjct: 244 VWKWRNNMVFED---SPWSVDEAWRKLGHEHDEFITFSDGDA--------GDPGSWLSSR 390
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYL 93
W PP +K+N DGS GG++RD G +L
Sbjct: 391 WQPPRQGTVKLNVDGSGREQDRCMGAGGLIRDDQGKWL 504
>BI418523
Length = 496
Score = 42.0 bits (97), Expect = 9e-05
Identities = 29/86 (33%), Positives = 38/86 (43%)
Frame = +1
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W PP+ +IK N DG+ + GGV RD N L FS G A++ I +
Sbjct: 187 WSPPSAGFIKCNVDGASQGNPGPSGVGGVFRDANRKILGYFSLNSGNGWAYEAKVRSILN 366
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLV 141
K ++IESDS AV V
Sbjct: 367 ALVFIQKFLLKNILIESDSTVAVEWV 444
>BP075797
Length = 504
Score = 38.1 bits (87), Expect = 0.001
Identities = 30/104 (28%), Positives = 46/104 (43%), Gaps = 1/104 (0%)
Frame = -2
Query: 84 VLRDHNGNYLVGFSA-LLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVN 142
+ +H G ++ S LL AE + L D F+ + +ESD V+ +
Sbjct: 473 IFYNHEGKAIIASSFHLLVAYEPAIAEALSLRRTITLPQDENFRIIELESDCLQLVHAWD 294
Query: 143 KGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAK 186
+ A++V++ K S D LKH R+IN VAD AK
Sbjct: 293 RSVPHPHYLASIVQDCKDLSSFSDSCSLKHSSRKINEVADHLAK 162
>TC11605
Length = 546
Score = 35.4 bits (80), Expect = 0.008
Identities = 21/65 (32%), Positives = 29/65 (44%)
Frame = +3
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W PP + +K N DGS ++ GG+LR+ L FS LG AE+ I
Sbjct: 348 WCPPEANILKFNVDGSARGSPGISGAGGILRNAESAVLGRFSKPLGVLCAYQAEVKAILL 527
Query: 116 GAKLA 120
K +
Sbjct: 528 ALKFS 542
>TC9793
Length = 568
Score = 35.0 bits (79), Expect = 0.010
Identities = 32/133 (24%), Positives = 59/133 (44%), Gaps = 2/133 (1%)
Frame = +2
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSAL-LGRCSINFAELWGIY 114
W PP + IK+N D +S S V R HN LV + L + AE +
Sbjct: 62 WEPPPEGIIKVNIDAGWSG-SCSTGFSLVARSHNAVMLVAATHLEETQIEPTLAEALALR 238
Query: 115 HGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTL-SNFDQVHLKHV 173
+ + +++++ES S +N +++ +P LV R L + F+ ++ KH+
Sbjct: 239 WCLSMIKEMEMERIIVESYSLVVLNTLHQ--KTKRPDIELVVYDSRELAAEFNFINFKHI 412
Query: 174 FREINSVADAFAK 186
R++ + F +
Sbjct: 413 GRKVINSGTCFGR 451
>BP067210
Length = 473
Score = 33.5 bits (75), Expect = 0.030
Identities = 20/60 (33%), Positives = 32/60 (53%)
Frame = -2
Query: 154 LVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDS 213
L+ +I+ L+ +VH+ H+ RE + VADAFAK + + A S + L HD+
Sbjct: 271 LISSIREMLAWNWRVHIIHIDRERSRVADAFAKQVARMRCPLHVLKAPHSNVISFLCHDT 92
>CB827358
Length = 453
Score = 32.7 bits (73), Expect = 0.052
Identities = 21/89 (23%), Positives = 41/89 (45%)
Frame = +2
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGTGDLRWVPPND 61
+W A N+L+F+++ + +SQ + T+ + + + D W P
Sbjct: 194 LWEAHNKLIFQEVPFRCEAAIQRATSMSQVDVSTVDPSANGPV-------HDAIWRRPPR 352
Query: 62 DWIKINTDGSYSPVSNLAACGGVLRDHNG 90
K+N DGS+ +++ G V R+H+G
Sbjct: 353 GVFKVNFDGSWKS-DSVSGIGMVARNHDG 436
>BP033684
Length = 557
Score = 32.3 bits (72), Expect = 0.068
Identities = 19/56 (33%), Positives = 25/56 (43%)
Frame = -1
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELW 111
W PP+ +K N DG+ + GGVL D+ S +LG SIN W
Sbjct: 287 WTPPSRGILKFNVDGASQGNPGPSGVGGVLCDYR-------SMVLGFFSINMGHGW 141
>AV780577
Length = 486
Score = 31.6 bits (70), Expect = 0.12
Identities = 25/84 (29%), Positives = 36/84 (42%), Gaps = 1/84 (1%)
Frame = +2
Query: 52 GDLRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLV-GFSALLGRCSINFAEL 110
G W PP +KIN P ++ G V+RD NG LV G S S+ E
Sbjct: 224 GPSTWWPPPHGKLKINIVDVVVPQVDMMGFGFVMRDANGLVLVSGASKHRRSLSVAMGEA 403
Query: 111 WGIYHGAKLALDRGFKKVMIESDS 134
+++ D G + +M S+S
Sbjct: 404 LA*RWAMQVSFDLGHRVLMFVSNS 475
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,195,380
Number of Sequences: 28460
Number of extensions: 54482
Number of successful extensions: 231
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 226
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 227
length of query: 224
length of database: 4,897,600
effective HSP length: 87
effective length of query: 137
effective length of database: 2,421,580
effective search space: 331756460
effective search space used: 331756460
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0010.5


