

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
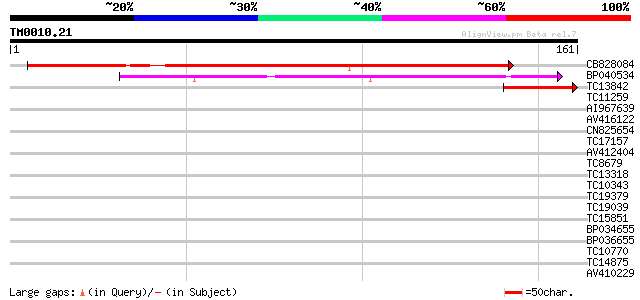
Query= TM0010.21
(161 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB828084 210 8e-56
BP040534 72 5e-14
TC13842 47 1e-06
TC11259 similar to UP|BAC98492 (BAC98492) AG-motif binding prote... 35 0.005
AI967639 34 0.011
AV416122 30 0.15
CN825654 30 0.20
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 30 0.26
AV412404 29 0.34
TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partia... 29 0.34
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 29 0.45
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 28 0.58
TC19379 homologue to UP|Q945P0 (Q945P0) Transcriptional activato... 28 0.58
TC19039 similar to UP|Q8L7W5 (Q8L7W5) AT4g12110/F16J13_180, part... 28 0.76
TC15851 28 0.76
BP034655 28 0.99
BP036655 28 0.99
TC10770 weakly similar to UP|RN12_HUMAN (Q9NVW2) RING finger pro... 28 0.99
TC14875 similar to UP|Q9VNX6 (Q9VNX6) CG7421-PA, partial (4%) 27 1.3
AV410229 27 1.3
>CB828084
Length = 528
Score = 210 bits (535), Expect = 8e-56
Identities = 112/139 (80%), Positives = 125/139 (89%), Gaps = 1/139 (0%)
Frame = +1
Query: 6 LFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKEL 65
L RRLAPSLAARFRQN++ +FSSSSSS VLH AT SSS+VEPVH+TENC+RRMKEL
Sbjct: 1 LIRRLAPSLAARFRQNERFLNFSSSSSS-VLHHAT----SSSYVEPVHITENCVRRMKEL 165
Query: 66 EASESATAPKTLRLSVETGGCSGFQYAFDL-DDRINSDDRVFEREGIKLVVDNISYDFVK 124
EASES T+ K LRLSVETGGCSGFQYAFDL DD +NSDDRVFE+EGIKLVVD++SYDFVK
Sbjct: 166 EASESTTSRKMLRLSVETGGCSGFQYAFDLDDDSVNSDDRVFEKEGIKLVVDSVSYDFVK 345
Query: 125 GATVDYVEELIRSAFVVTE 143
GATVDYVEELIRSAFVV++
Sbjct: 346 GATVDYVEELIRSAFVVSQ 402
>BP040534
Length = 577
Score = 72.0 bits (175), Expect = 5e-14
Identities = 44/129 (34%), Positives = 68/129 (52%), Gaps = 3/129 (2%)
Frame = -3
Query: 32 SSSVLHDATTSSSSSSHVEP-VHMTENCLRRMKELEASESATAPKTLRLSVETGGCSGFQ 90
S S+ + ++ SS V P V +T+ L+ + + + ++ LR+ V+ GGCSG
Sbjct: 575 SLSIRSVSVPAAPSSGSVAPAVSLTDQALKHLNRMRSEKNQDL--CLRIGVKQGGCSGMS 402
Query: 91 YAFDLDDRINS--DDRVFEREGIKLVVDNISYDFVKGATVDYVEELIRSAFVVTENPSAV 148
Y+ D +DR N+ DD + E EG +V D S F+ G +DY + LI F +NP+A
Sbjct: 401 YSMDFEDRANTRPDDSIIEYEGFVIVCDPKSLLFIFGMQLDYSDALIGGGFSF-KNPNAT 225
Query: 149 GGCSCKSSF 157
C C SF
Sbjct: 224 QTCGCGKSF 198
>TC13842
Length = 412
Score = 47.4 bits (111), Expect = 1e-06
Identities = 21/21 (100%), Positives = 21/21 (100%)
Frame = +3
Query: 141 VTENPSAVGGCSCKSSFMVKQ 161
VTENPSAVGGCSCKSSFMVKQ
Sbjct: 180 VTENPSAVGGCSCKSSFMVKQ 242
>TC11259 similar to UP|BAC98492 (BAC98492) AG-motif binding protein-2,
partial (12%)
Length = 468
Score = 35.4 bits (80), Expect = 0.005
Identities = 25/74 (33%), Positives = 36/74 (47%), Gaps = 1/74 (1%)
Frame = -2
Query: 13 SLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMK-ELEASESA 71
SL + + L FSSSSSSS + D SS+ P+ + C R++ A+ +A
Sbjct: 236 SLHIGLSEGKALLGFSSSSSSSPMSDEKFKRSSTEPRAPITIPAGCWLRLE*AAAAAAAA 57
Query: 72 TAPKTLRLSVETGG 85
+ R SVE GG
Sbjct: 56 RRRRNPRKSVERGG 15
>AI967639
Length = 335
Score = 34.3 bits (77), Expect = 0.011
Identities = 27/70 (38%), Positives = 39/70 (55%)
Frame = -1
Query: 8 RRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEA 67
R L+ SL++ ++ +S SSSSSSS L+ + SSSSSS P + E L ++ L
Sbjct: 227 RGLSLSLSSLMLKSSSSSSSSSSSSSSRLYSSP*SSSSSSS-SPSELLEISLLCLRFLRY 51
Query: 68 SESATAPKTL 77
S S T T+
Sbjct: 50 SSSYTTSPTI 21
>AV416122
Length = 414
Score = 30.4 bits (67), Expect = 0.15
Identities = 30/83 (36%), Positives = 39/83 (46%), Gaps = 7/83 (8%)
Frame = -3
Query: 18 FRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMT--ENCLRRMK-----ELEASES 70
FR SSSSSSS ++ SSSSSS P + T L + E ++S +
Sbjct: 400 FRSTSASPFESSSSSSSSSSSSSLSSSSSSSSSPSNWTSLSGSLSSLAASLGWEEQSSVA 221
Query: 71 ATAPKTLRLSVETGGCSGFQYAF 93
TA L LS E+G SG +F
Sbjct: 220 ETAGWLLLLSFESGLGSGVAASF 152
>CN825654
Length = 580
Score = 30.0 bits (66), Expect = 0.20
Identities = 17/25 (68%), Positives = 20/25 (80%)
Frame = -2
Query: 23 KLTSFSSSSSSSVLHDATTSSSSSS 47
KLT+ SSSSSS V D ++SSSSSS
Sbjct: 144 KLTTCSSSSSSPVNSDFSSSSSSSS 70
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 29.6 bits (65), Expect = 0.26
Identities = 19/40 (47%), Positives = 23/40 (57%)
Frame = -3
Query: 8 RRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
R L P L F +S SSSSSSS +++SSSSSS
Sbjct: 912 RALLPLLLLLFPSPSSSSSSSSSSSSSSSSSSSSSSSSSS 793
Score = 26.9 bits (58), Expect = 1.7
Identities = 17/38 (44%), Positives = 23/38 (59%)
Frame = -3
Query: 10 LAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
L PS ++ + +S SSSSSSS ++ SSSSSS
Sbjct: 885 LFPSPSSSSSSSSSSSSSSSSSSSSSSSSSSPSSSSSS 772
Score = 26.6 bits (57), Expect = 2.2
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVEP 51
+S SSSSSSS +++SSSSSS P
Sbjct: 870 SSSSSSSSSSSSSSSSSSSSSSSSSSP 790
Score = 24.6 bits (52), Expect = 8.4
Identities = 15/37 (40%), Positives = 22/37 (58%)
Frame = -3
Query: 11 APSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
+PS ++ + +S SSSSSSS ++SSSS S
Sbjct: 876 SPSSSSSSSSSSSSSSSSSSSSSSSSSSPSSSSSSIS 766
>AV412404
Length = 386
Score = 29.3 bits (64), Expect = 0.34
Identities = 21/58 (36%), Positives = 32/58 (54%)
Frame = +2
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVE 82
+SFSSSSSS + +TT++ S H T AS+++++P +L LSVE
Sbjct: 65 SSFSSSSSSLSIPKSTTTTLSLLSFRRNHST--------PAAASKTSSSPSSLPLSVE 214
>TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partial (70%)
Length = 972
Score = 29.3 bits (64), Expect = 0.34
Identities = 18/32 (56%), Positives = 23/32 (71%), Gaps = 3/32 (9%)
Frame = -1
Query: 24 LTSFSSSSSSSVLHDATTSSSSS---SHVEPV 52
L S+SSSSSSS ++TSSSSS SH +P+
Sbjct: 714 LLSYSSSSSSSSSTSSSTSSSSSSSPSHPKPL 619
Score = 28.5 bits (62), Expect = 0.58
Identities = 17/32 (53%), Positives = 21/32 (65%)
Frame = -1
Query: 24 LTSFSSSSSSSVLHDATTSSSSSSHVEPVHMT 55
L+ SSSSSSS +T+SSSSSS P +T
Sbjct: 711 LSYSSSSSSSSSTSSSTSSSSSSSPSHPKPLT 616
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 28.9 bits (63), Expect = 0.45
Identities = 18/38 (47%), Positives = 24/38 (62%)
Frame = -3
Query: 10 LAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
LAPS + + +S SSSSSSS +++SSSSSS
Sbjct: 415 LAPSFPSTSTSSLPSSSSSSSSSSSSSSPSSSSSSSSS 302
Score = 28.9 bits (63), Expect = 0.45
Identities = 20/59 (33%), Positives = 33/59 (55%)
Frame = -3
Query: 26 SFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVETG 84
SF S+S+SS+ +++SSSSSS P + + +S S+++ + LSV TG
Sbjct: 406 SFPSTSTSSLPSSSSSSSSSSSSSSP-SSSSSSSSSSSSSSSSSSSSSSSSSSLSV*TG 233
Score = 27.7 bits (60), Expect = 0.99
Identities = 19/45 (42%), Positives = 27/45 (59%)
Frame = -3
Query: 3 TSSLFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
TSSL + S ++ + +S SSSSSSS +++SSSSSS
Sbjct: 388 TSSLPSSSSSSSSSSSSSSPSSSSSSSSSSSSSSSSSSSSSSSSS 254
Score = 26.6 bits (57), Expect = 2.2
Identities = 16/36 (44%), Positives = 22/36 (60%)
Frame = -3
Query: 12 PSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
PS + + +S SSSSSSS +++SSSSSS
Sbjct: 400 PSTSTSSLPSSSSSSSSSSSSSSPSSSSSSSSSSSS 293
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 319 SSSSSSSSSSSSSSSSSSSSSSS 251
Score = 24.6 bits (52), Expect = 8.4
Identities = 23/78 (29%), Positives = 35/78 (44%)
Frame = -3
Query: 11 APSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASES 70
A LA F + SSSSSSS +++ SSSSS + + +S S
Sbjct: 424 AAPLAPSFPSTSTSSLPSSSSSSSSSSSSSSPSSSSSSSSSSSSSSS--------SSSSS 269
Query: 71 ATAPKTLRLSVETGGCSG 88
+++ +L + GG SG
Sbjct: 268 SSSSSSLSV*TGLGGPSG 215
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 28.5 bits (62), Expect = 0.58
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 1/64 (1%)
Frame = -1
Query: 14 LAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESA-T 72
++ R N + SSSS+++ AT +SSSSS +CL+R +S S+ T
Sbjct: 264 MSVRLNNNSAPSLHSSSSATTASSSATPASSSSS------TATSCLKRGTSTSSSSSSIT 103
Query: 73 APKT 76
+P +
Sbjct: 102 SPSS 91
>TC19379 homologue to UP|Q945P0 (Q945P0) Transcriptional activator FHA1,
partial (56%)
Length = 525
Score = 28.5 bits (62), Expect = 0.58
Identities = 23/76 (30%), Positives = 38/76 (49%)
Frame = +1
Query: 2 QTSSLFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRR 61
+TSS R PS +A +N TS S +S+++ ATT +SS++ P + +
Sbjct: 85 RTSSTTCRPTPSSSAATPRNPPSTSTSPASAAA*TSPATTPASSTT--SPAADSPSRFSA 258
Query: 62 MKELEASESATAPKTL 77
+ S T+P+TL
Sbjct: 259 RTAASSRASFTSPETL 306
>TC19039 similar to UP|Q8L7W5 (Q8L7W5) AT4g12110/F16J13_180, partial (61%)
Length = 763
Score = 28.1 bits (61), Expect = 0.76
Identities = 20/50 (40%), Positives = 26/50 (52%)
Frame = +2
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAP 74
T+FSSSSSSS + +SSS S+ P +T + R SAT P
Sbjct: 269 TTFSSSSSSSPSSLSPSSSSRSNASPPSTITRSNPRFASPSVTCSSATRP 418
>TC15851
Length = 684
Score = 28.1 bits (61), Expect = 0.76
Identities = 18/60 (30%), Positives = 24/60 (40%), Gaps = 3/60 (5%)
Frame = -2
Query: 35 VLHDATTSSSSSSHVEPVHMTENCLRR---MKELEASESATAPKTLRLSVETGGCSGFQY 91
+ H T SS S H + C + + ASE + +P L TG CSG Y
Sbjct: 182 ISHPITLSSQESKHFSVTGNSTVCFAAGLIVISVTASEFSNSPSLLSPPYLTGSCSGAMY 3
>BP034655
Length = 517
Score = 27.7 bits (60), Expect = 0.99
Identities = 16/34 (47%), Positives = 20/34 (58%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENC 58
+S SSSSSSS +++SSSSSS P C
Sbjct: 206 SSSSSSSSSSSSSSSSSSSSSSSSESPKSAAVRC 105
Score = 25.8 bits (55), Expect = 3.8
Identities = 15/26 (57%), Positives = 19/26 (72%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSSHVE 50
+S SSSSSSS +++SSSSSS E
Sbjct: 209 SSSSSSSSSSSSSSSSSSSSSSSSSE 132
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 248 SSSSSSSSSSSSSSSSSSSSSSS 180
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 242 SSSSSSSSSSSSSSSSSSSSSSS 174
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 239 SSSSSSSSSSSSSSSSSSSSSSS 171
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 236 SSSSSSSSSSSSSSSSSSSSSSS 168
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 230 SSSSSSSSSSSSSSSSSSSSSSS 162
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 233 SSSSSSSSSSSSSSSSSSSSSSS 165
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 227 SSSSSSSSSSSSSSSSSSSSSSS 159
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 224 SSSSSSSSSSSSSSSSSSSSSSS 156
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 221 SSSSSSSSSSSSSSSSSSSSSSS 153
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 218 SSSSSSSSSSSSSSSSSSSSSSS 150
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 215 SSSSSSSSSSSSSSSSSSSSSSS 147
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 212 SSSSSSSSSSSSSSSSSSSSSSS 144
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 245 SSSSSSSSSSSSSSSSSSSSSSS 177
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 251 SSSSSSSSSSSSSSSSSSSSSSS 183
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/23 (60%), Positives = 18/23 (77%)
Frame = -3
Query: 25 TSFSSSSSSSVLHDATTSSSSSS 47
+S SSSSSSS +++SSSSSS
Sbjct: 254 SSSSSSSSSSSSSSSSSSSSSSS 186
>BP036655
Length = 556
Score = 27.7 bits (60), Expect = 0.99
Identities = 17/45 (37%), Positives = 25/45 (54%)
Frame = +1
Query: 3 TSSLFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSS 47
+SS + S++ + + SS SSS LH +TTSSSSS+
Sbjct: 235 SSSSSSSFSVSISVSLSSSTSSSLSSSLISSSSLHSSTTSSSSST 369
Score = 25.0 bits (53), Expect = 6.4
Identities = 19/47 (40%), Positives = 25/47 (52%)
Frame = +1
Query: 5 SLFRRLAPSLAARFRQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEP 51
SL + SL++ + L S ++SSSSS TTSSSS S P
Sbjct: 277 SLSSSTSSSLSSSLISSSSLHSSTTSSSSS-----TTSSSSPSSERP 402
>TC10770 weakly similar to UP|RN12_HUMAN (Q9NVW2) RING finger protein 12
(LIM domain interacting RING finger protein) (RING
finger LIM domain-binding protein) (R-LIM) (NY-REN-43
antigen), partial (4%)
Length = 481
Score = 27.7 bits (60), Expect = 0.99
Identities = 25/89 (28%), Positives = 42/89 (47%)
Frame = -2
Query: 19 RQNQKLTSFSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLR 78
+QNQ+ F +S++ ++ +SSSSSS P + + R E +S S T
Sbjct: 345 QQNQRFIKFYIRTSAAFVYSIMSSSSSSS--SPSSRSASSSSRGTEGSSSPSVTD----- 187
Query: 79 LSVETGGCSGFQYAFDLDDRINSDDRVFE 107
++ G S F+Y+ L+ R R F+
Sbjct: 186 -AISDGWYSLFRYSDMLNLRSGFSSRSFK 103
>TC14875 similar to UP|Q9VNX6 (Q9VNX6) CG7421-PA, partial (4%)
Length = 1240
Score = 27.3 bits (59), Expect = 1.3
Identities = 17/59 (28%), Positives = 30/59 (50%)
Frame = -3
Query: 27 FSSSSSSSVLHDATTSSSSSSHVEPVHMTENCLRRMKELEASESATAPKTLRLSVETGG 85
F S+ SSS+ HD++ S +H P+ + R ++L++S A + S+ T G
Sbjct: 785 FHSTHSSSMPHDSSALSPGENHSPPLSPAPS---RTRDLKSSGLAARGSSGHRSLRTSG 618
>AV410229
Length = 345
Score = 27.3 bits (59), Expect = 1.3
Identities = 13/28 (46%), Positives = 20/28 (71%)
Frame = -1
Query: 4 SSLFRRLAPSLAARFRQNQKLTSFSSSS 31
SS+FRR++ S+A + KL+SFS +S
Sbjct: 105 SSIFRRVSASIAKKPNLTLKLSSFSKNS 22
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.126 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,682,064
Number of Sequences: 28460
Number of extensions: 43877
Number of successful extensions: 988
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 553
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 773
length of query: 161
length of database: 4,897,600
effective HSP length: 83
effective length of query: 78
effective length of database: 2,535,420
effective search space: 197762760
effective search space used: 197762760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0010.21


