

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.2
(370 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
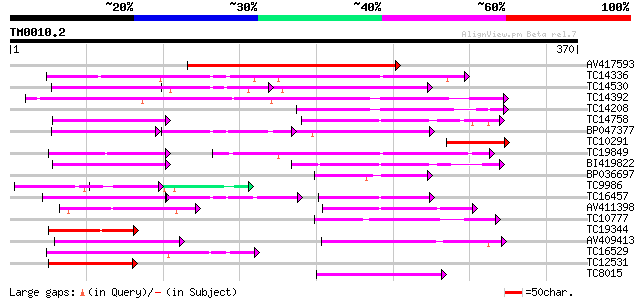
Score E
Sequences producing significant alignments: (bits) Value
AV417593 213 4e-56
TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 87 4e-18
TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribos... 72 1e-13
TC14392 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, pa... 69 1e-12
TC14208 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, pa... 59 1e-09
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 53 7e-08
BP047377 53 9e-08
TC10291 similar to UP|Q9ASP6 (Q9ASP6) AT4g00830/A_TM018A10_14, p... 52 1e-07
TC19849 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, p... 50 6e-07
BI419822 49 1e-06
BP036697 48 2e-06
TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding prot... 47 5e-06
TC16457 similar to UP|Q41124 (Q41124) Chloroplast RNA binding pr... 47 7e-06
AV411398 46 9e-06
TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor S... 46 1e-05
TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG polyaden... 45 1e-05
AV409413 45 2e-05
TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 45 2e-05
TC12531 similar to UP|Q9ASQ8 (Q9ASQ8) At1g22910/F19G10_13, parti... 44 3e-05
TC8015 similar to UP|ROC4_NICSY (P19683) 31 kDa ribonucleoprotei... 44 4e-05
>AV417593
Length = 423
Score = 213 bits (542), Expect = 4e-56
Identities = 96/139 (69%), Positives = 122/139 (87%)
Frame = +2
Query: 117 WTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKL 176
W ED++K V + GPGV ++EL+KD+++ NRGFAFI+YYNHACAE+SRQKM + FKL
Sbjct: 5 WGEEDLRKTVTEAGPGVTAVELVKDVKNINNNRGFAFIDYYNHACAEHSRQKMMSPTFKL 184
Query: 177 ENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKS 236
+NAPTVSWA+P+N++SSA SQVKAVYVKNLP+N+TQ+ ++KLFEHHGKITKVVLPP KS
Sbjct: 185 GDNAPTVSWADPKNADSSASSQVKAVYVKNLPKNVTQEQVRKLFEHHGKITKVVLPPPKS 364
Query: 237 GQEKSRFGFVHFSDRSSAM 255
GQEK+R GFVHF++RS+AM
Sbjct: 365 GQEKNRIGFVHFAERSNAM 421
>TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(85%)
Length = 1917
Score = 87.0 bits (214), Expect = 4e-18
Identities = 74/302 (24%), Positives = 137/302 (44%), Gaps = 26/302 (8%)
Frame = +2
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
+ ++I + + + L + G + ++ SG+ KGY FV F+T+E A AI
Sbjct: 89 AGNIFIKNLDKAIDHKALHDTFSTFGNILSCKVA-TDSSGQSKGYGFVQFETEEAAQNAI 265
Query: 85 EELNNSEFKGKKI-----------KCSSSQAK-HRLFIGNIPKKWTVEDMKKVVADVGPG 132
E+LN K++ + S+ +AK + +F+ N+ + T +++K V + G
Sbjct: 266 EKLNGMLLNDKQVYVGPFLRKQERESSTDKAKFNNVFVKNLSESTTDDELKNVFGEFGT- 442
Query: 133 VISIELLKDLQSSGRNRGFAFIEYYN-----HACAEYSRQKMSNSNF-------KLENNA 180
+ S +++D G+++ F F+ + N A + +K+ + + K E
Sbjct: 443 ITSAVVMRD--GDGKSKCFGFVNFENADDAARAVESLNGKKVDDKEWYVGKAQKKSEREQ 616
Query: 181 PTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEK 240
+ E E++ Q +YVKNL ++I + LK+LF +G IT + +G +
Sbjct: 617 ELKNKFEQSMKEAADKYQGANLYVKNLDDSIADEKLKELFSSYGTITSCKVMRDPNGISR 796
Query: 241 SRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRS--SGTSNSQKPVVLPT 298
GFV FS A +AL + K L +LA+ + D+R+ +PV +P
Sbjct: 797 GS-GFVAFSTPEEASRALLEMNGKMVVSKPLYVTLAQRKEDRRARLQAQFAQMRPVAMPP 973
Query: 299 YP 300
P
Sbjct: 974 AP 979
>TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribosomal
protein 2, chloroplast precursor (PSRP-2), partial (68%)
Length = 1104
Score = 72.4 bits (176), Expect = 1e-13
Identities = 52/185 (28%), Positives = 86/185 (46%), Gaps = 8/185 (4%)
Frame = +2
Query: 100 SSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFI----- 154
+ S+A RL++GNIP+ T +++ +V + V E++ D + SGR+R FAF+
Sbjct: 212 TQSEADRRLYVGNIPRTLTNDELANIVQEHA-AVEKAEVMYD-KYSGRSRRFAFVTVKTV 385
Query: 155 EYYNHACAEYSRQKMSNSNFKL---ENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENI 211
E N A + + ++ K+ E TV + + ES V VYV NL + +
Sbjct: 386 EDANAAIEKLNGTQIGGREIKVNVTEKPLTTVDFPLVQAEESEYVDSPYKVYVGNLAKTV 565
Query: 212 TQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNL 271
T D LK F GK+ + + S FGFV FS A+ + ++G+ +
Sbjct: 566 TSDMLKNFFSEKGKVLSAKVSRVPGTSKSSGFGFVTFSSDEDVEAAISSFNNAPLEGQKI 745
Query: 272 ECSLA 276
+ A
Sbjct: 746 RVNKA 760
Score = 59.7 bits (143), Expect = 8e-10
Identities = 38/165 (23%), Positives = 82/165 (49%), Gaps = 20/165 (12%)
Frame = +2
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
+Y+G I ++ ++L Q V + +M K SG + +AFV KT E A+ AIE+L
Sbjct: 236 LYVGNIPRTLTNDELANIVQEHAAVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKL 415
Query: 88 NNSEFKGKKIKCSSSQ--------------------AKHRLFIGNIPKKWTVEDMKKVVA 127
N ++ G++IK + ++ + +++++GN+ K T + +K +
Sbjct: 416 NGTQIGGREIKVNVTEKPLTTVDFPLVQAEESEYVDSPYKVYVGNLAKTVTSDMLKNFFS 595
Query: 128 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNS 172
+ G V+S ++ + + + ++ GF F+ + + E + +N+
Sbjct: 596 EKGK-VLSAKVSR-VPGTSKSSGFGFVTFSSDEDVEAAISSFNNA 724
>TC14392 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, partial (77%)
Length = 1576
Score = 68.9 bits (167), Expect = 1e-12
Identities = 68/345 (19%), Positives = 141/345 (40%), Gaps = 30/345 (8%)
Frame = +3
Query: 11 EKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYA 70
++ +HA+ + P ++IG + + E L GE++ V++++ K + + +GY
Sbjct: 291 QQHQHAQP-STPDEVRTLWIGDLQYWMDENYLYQCFAHTGELATVKVIRNKNTNQSEGYG 467
Query: 71 FVAFKTKELASQAIE--------------ELNNSEFKGKKIKCSSSQAKHRLFIGNIPKK 116
F+ F ++ A + ++ LN + F + + +F+G++
Sbjct: 468 FLEFTSRAGAERILQTYNGTIMPNGGQNFRLNWATFSAGGERRHDDSPDYTIFVGDLAAD 647
Query: 117 WTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKM------- 169
T + +V V +++ D +S R +G+ F+ + + + + +M
Sbjct: 648 VTDYHLTEVFRTRYNSVKGAKVVIDRLTS-RTKGYGFVRFADESEQVRAMTEMQGVVCST 824
Query: 170 --------SNSNFKLENNA-PTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLF 220
+N N P S+ + ++ ++V NL N+T D L+++F
Sbjct: 825 RPMRIGPAANKNLGTGTGTQPKASYQNSQGPQNENDPNNTTIFVGNLDPNVTDDHLRQVF 1004
Query: 221 EHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEA 280
+G + V +P K R GFV F+DRS A +AL+ + G+N+ S + +
Sbjct: 1005GLYGDLVHVKIPQGK------RCGFVQFADRSCAEEALRVLNGTLLGGQNVRLSWGRSPS 1166
Query: 281 DQRSSGTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGYGAAGFA 325
++++ P + G Y G GY G+A
Sbjct: 1167NKQTQSD------------PSQWNNSAGGAGYYGYAQGYENYGYA 1265
>TC14208 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, partial (41%)
Length = 837
Score = 59.3 bits (142), Expect = 1e-09
Identities = 39/138 (28%), Positives = 65/138 (46%)
Frame = +2
Query: 188 PRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVH 247
P+ S++ ++V NL N+ D L+++F +G++ V +P K R GFV
Sbjct: 155 PQGSQNENDPSNTTIFVGNLDPNVNDDHLRQVFSPYGELVHVKIPAGK------RCGFVQ 316
Query: 248 FSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGM 307
F+DRS A +AL+ + G+N+ S + +++++ SN Y
Sbjct: 317 FADRSGAEEALRVLNGTLLGGQNVRLSWGRSPSNKQAQPDSNQ--------------YAA 454
Query: 308 VGGAYGGIGAGYGAAGFA 325
GG Y G G GY G+A
Sbjct: 455 AGGYY-GYGQGYENYGYA 505
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 53.1 bits (126), Expect = 7e-08
Identities = 23/77 (29%), Positives = 43/77 (54%)
Frame = +1
Query: 29 YIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEELN 88
++GG+A L S GE+ + +++ +E+G +G+ FV F ++E AIE +N
Sbjct: 91 FVGGLAWTTDSHTLEQAFSSYGEIIDSKVVNDRETGRSRGFGFVTFTSEEAMRSAIEGMN 270
Query: 89 NSEFKGKKIKCSSSQAK 105
+E G+ I + +QA+
Sbjct: 271 GNELDGRNITVNEAQAR 321
Score = 40.4 bits (93), Expect = 5e-04
Identities = 38/141 (26%), Positives = 61/141 (42%), Gaps = 8/141 (5%)
Frame = +1
Query: 191 SESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKIT-KVVLPPAKSGQEKSRFGFVHFS 249
S+S+ +V L +L++ F +G+I V+ ++G+ + FGFV F+
Sbjct: 55 SDSAMADVEYRCFVGGLAWTTDSHTLEQAFSSYGEIIDSKVVNDRETGRSRG-FGFVTFT 231
Query: 250 DRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYP-----HRLG 304
+ A++ E+DG+N+ + EA R G + Y G
Sbjct: 232 SEEAMRSAIEGMNGNELDGRNITVN----EAQARGGGGGGGGRGG--GGYGGGGGRREGG 393
Query: 305 YGMVGGA--YGGIGAGYGAAG 323
YG GG+ YGG G GYG G
Sbjct: 394 YGGGGGSRGYGGGGGGYGGGG 456
>BP047377
Length = 564
Score = 52.8 bits (125), Expect = 9e-08
Identities = 25/71 (35%), Positives = 40/71 (56%)
Frame = +3
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
V++G I + +EE L CQ VG V R++ +E+G+ KGY F +K +E A A L
Sbjct: 135 VFVGNIPYDATEEQLIEICQEVGPVVSFRLVIDRETGKPKGYGFCEYKDEETALSARRNL 314
Query: 88 NNSEFKGKKIK 98
E G++++
Sbjct: 315 QGYEINGRQLR 347
Score = 49.3 bits (116), Expect = 1e-06
Identities = 27/100 (27%), Positives = 50/100 (50%), Gaps = 7/100 (7%)
Frame = +3
Query: 185 WAEPRNSESSAV-------SQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSG 237
W+ + SSAV SQ + V+V N+P + T++ L ++ + G + L +
Sbjct: 63 WSSRASPSSSAVDFMASSQSQHRCVFVGNIPYDATEEQLIEICQEVGPVVSFRLVIDRET 242
Query: 238 QEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAK 277
+ +GF + D +A+ A +N + YEI+G+ L A+
Sbjct: 243 GKPKGYGFCEYKDEETALSARRNLQGYEINGRQLRVDFAE 362
Score = 47.0 bits (110), Expect = 5e-06
Identities = 29/89 (32%), Positives = 53/89 (58%), Gaps = 1/89 (1%)
Frame = +3
Query: 100 SSSQAKHR-LFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYN 158
+SSQ++HR +F+GNIP T E + ++ +VGP V+S L+ D + +G+ +G+ F EY +
Sbjct: 108 ASSQSQHRCVFVGNIPYDATEEQLIEICQEVGP-VVSFRLVID-RETGKPKGYGFCEYKD 281
Query: 159 HACAEYSRQKMSNSNFKLENNAPTVSWAE 187
A +R+ + +++ V +AE
Sbjct: 282 EETALSARRNL--QGYEINGRQLRVDFAE 362
>TC10291 similar to UP|Q9ASP6 (Q9ASP6) AT4g00830/A_TM018A10_14, partial (7%)
Length = 674
Score = 52.4 bits (124), Expect = 1e-07
Identities = 22/41 (53%), Positives = 29/41 (70%)
Frame = +3
Query: 286 GTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGYGAAGFAQ 326
G ++QK +LP YP +GYG++G YG +GAGYGA G AQ
Sbjct: 15 GVLHTQKQGLLPRYPPSVGYGLLGAPYGALGAGYGAPGLAQ 137
>TC19849 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, partial
(30%)
Length = 603
Score = 50.1 bits (118), Expect = 6e-07
Identities = 47/196 (23%), Positives = 82/196 (40%), Gaps = 12/196 (6%)
Frame = +2
Query: 133 VISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNF------------KLENNA 180
+ S +++D+ G+++ F F+ + N A + ++ F K E
Sbjct: 5 ITSAVVMRDVD--GKSKCFGFVNFENADEAAKAVDALNGKKFDDKEWYVGKAKKKSEREL 178
Query: 181 PTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEK 240
E E+ Q +Y+KNL +N+ + L++LF G IT + G +
Sbjct: 179 ELKEQHEQSMQETVDKYQGANLYLKNLDDNVDDEKLRELFSEFGTITSCKIMRDPHGVSR 358
Query: 241 SRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYP 300
GFV FS A +AL +DGK L +LA+ + ++R+ + +LP
Sbjct: 359 GS-GFVAFSTPEEATRALGEMNGKMVDGKPLYVALAQKKEERRAKLQGGYFQQQLLPGM- 532
Query: 301 HRLGYGMVGGAYGGIG 316
R G G + Y +G
Sbjct: 533 -RPGGGPLSNFYVPLG 577
Score = 46.2 bits (108), Expect = 9e-06
Identities = 25/80 (31%), Positives = 43/80 (53%)
Frame = +2
Query: 26 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIE 85
+ +Y+ + +NV +E LR G ++ +IM+ G +G FVAF T E A++A+
Sbjct: 236 ANLYLKNLDDNVDDEKLRELFSEFGTITSCKIMRDPH-GVSRGSGFVAFSTPEEATRALG 412
Query: 86 ELNNSEFKGKKIKCSSSQAK 105
E+N GK + + +Q K
Sbjct: 413 EMNGKMVDGKPLYVALAQKK 472
>BI419822
Length = 380
Score = 49.3 bits (116), Expect = 1e-06
Identities = 21/77 (27%), Positives = 41/77 (52%)
Frame = +2
Query: 29 YIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEELN 88
++GG+A E L GE+ E +++ +E+G +G+ FV F +++ AIE +N
Sbjct: 86 FVGGLAWATDNEALEKAFSPYGEIVESKVINDRETGRSRGFGFVTFASEQAMKDAIEAMN 265
Query: 89 NSEFKGKKIKCSSSQAK 105
G+ I + +Q++
Sbjct: 266 GQNLDGRNITVNEAQSR 316
Score = 41.2 bits (95), Expect = 3e-04
Identities = 36/140 (25%), Positives = 60/140 (42%), Gaps = 1/140 (0%)
Frame = +2
Query: 185 WAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKV-VLPPAKSGQEKSRF 243
W +S +SA + + +V L ++L+K F +G+I + V+ ++G+ + F
Sbjct: 35 WFFSFSSMASAEIEFRC-FVGGLAWATDNEALEKAFSPYGEIVESKVINDRETGRSRG-F 208
Query: 244 GFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRL 303
GFV F+ + A++ +DG+N+ + EA R S
Sbjct: 209 GFVTFASEQAMKDAIEAMNGQNLDGRNITVN----EAQSRGSSGG--------------- 331
Query: 304 GYGMVGGAYGGIGAGYGAAG 323
GG YGG G GYG G
Sbjct: 332 -----GGRYGGGGGGYGGGG 376
>BP036697
Length = 578
Score = 48.1 bits (113), Expect = 2e-06
Identities = 28/79 (35%), Positives = 40/79 (50%), Gaps = 2/79 (2%)
Frame = +3
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVL--PPAKSGQEKSRFGFVHFSDRSSAMKA 257
+ VYV NLP +I + ++ LF +G IT + L PP G + FV F D A A
Sbjct: 96 RTVYVGNLPGDIREREVEDLFYKYGNITHIDLKVPPRPPG-----YAFVEFEDTQDAEDA 260
Query: 258 LKNTEKYEIDGKNLECSLA 276
++ + Y+ DG L LA
Sbjct: 261 IRGRDGYDFDGHRLRVELA 317
>TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (46%)
Length = 1150
Score = 47.0 bits (110), Expect = 5e-06
Identities = 31/101 (30%), Positives = 51/101 (49%), Gaps = 4/101 (3%)
Frame = +3
Query: 4 NHTNDEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQ----SVGEVSEVRIMK 59
N ++D + + E P + VY+G + ++V++ +L CQ G + EVRI +
Sbjct: 285 NGSSDGGQDNNNEEAPENNPSYTTVYVGNLPHDVTQAELH--CQFHALGAGVIEEVRIQR 458
Query: 60 AKESGERKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCS 100
K G+ FV + T E A+ AI+ N +GK +KCS
Sbjct: 459 DK------GFGFVRYNTHEEAALAIQVANGRIVRGKTMKCS 563
Score = 40.0 bits (92), Expect = 6e-04
Identities = 32/149 (21%), Positives = 59/149 (39%), Gaps = 42/149 (28%)
Frame = +3
Query: 53 SEVRIMKAKESGERKGYAFVAFKTKELASQAIEELNNSEFKGKKIKC---------SSSQ 103
S+ R+M ++G KGY FV+F+ + A +I ++ ++I+C SSS+
Sbjct: 66 SDARVMWDHKTGRSKGYGFVSFRDHQDAQSSINDMTGKWLGNRQIRCNWATKGAGGSSSE 245
Query: 104 AKH---------------------------------RLFIGNIPKKWTVEDMKKVVADVG 130
K+ +++GN+P T ++ +G
Sbjct: 246 EKNSDNQNAVVLTNGSSDGGQDNNNEEAPENNPSYTTVYVGNLPHDVTQAELHCQFHALG 425
Query: 131 PGVISIELLKDLQSSGRNRGFAFIEYYNH 159
GVI ++ R++GF F+ Y H
Sbjct: 426 AGVIEEVRIQ------RDKGFGFVRYNTH 494
Score = 39.3 bits (90), Expect = 0.001
Identities = 45/197 (22%), Positives = 73/197 (36%), Gaps = 36/197 (18%)
Frame = +3
Query: 145 SGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWA---------EPRNSESSA 195
+GR++G+ F+ + +H A+ S M+ L N +WA E +NS++
Sbjct: 96 TGRSKGYGFVSFRDHQDAQSSINDMTGK--WLGNRQIRCNWATKGAGGSSSEEKNSDNQN 269
Query: 196 V-----------------------SQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLP 232
VYV NLP ++TQ L F G V+
Sbjct: 270 AVVLTNGSSDGGQDNNNEEAPENNPSYTTVYVGNLPHDVTQAELHCQFHALGA---GVIE 440
Query: 233 PAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAK----PEADQRSSGTS 288
+ ++K FGFV ++ A A++ + GK ++CS P
Sbjct: 441 EVRIQRDKG-FGFVRYNTHEEAALAIQVANGRIVRGKTMKCSWGSRPTPPGTASNPLPPP 617
Query: 289 NSQKPVVLPTYPHRLGY 305
+Q +LPT GY
Sbjct: 618 AAQPYQILPTAGMNQGY 668
>TC16457 similar to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (30%)
Length = 655
Score = 46.6 bits (109), Expect = 7e-06
Identities = 23/84 (27%), Positives = 42/84 (49%)
Frame = +2
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P ++++G ++ V+ E L G V R++ E+G +GY FV + +
Sbjct: 2 PETEHKLFVGNLSWTVTSESLIQVFPEYGTVVGARVLYDGETGRSRGYGFVCYSKRSELE 181
Query: 82 QAIEELNNSEFKGKKIKCSSSQAK 105
A+ LNN E +G+ I+ S ++ K
Sbjct: 182 TALISLNNVELEGRAIRVSLAEGK 253
Score = 42.7 bits (99), Expect = 1e-04
Identities = 29/89 (32%), Positives = 48/89 (53%)
Frame = +2
Query: 103 QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACA 162
+ +H+LF+GN+ T E + +V + G V+ +L D +GR+RG+ F+ Y +
Sbjct: 5 ETEHKLFVGNLSWTVTSESLIQVFPEYGT-VVGARVLYD-GETGRSRGYGFVCYSKR--S 172
Query: 163 EYSRQKMSNSNFKLENNAPTVSWAEPRNS 191
E +S +N +LE A VS AE + S
Sbjct: 173 ELETALISLNNVELEGRAIRVSLAEGKRS 259
Score = 40.4 bits (93), Expect = 5e-04
Identities = 24/77 (31%), Positives = 44/77 (56%), Gaps = 1/77 (1%)
Frame = +2
Query: 202 VYVKNLPENITQDSLKKLFEHHGKITKV-VLPPAKSGQEKSRFGFVHFSDRSSAMKALKN 260
++V NL +T +SL ++F +G + VL ++G+ + +GFV +S RS AL +
Sbjct: 20 LFVGNLSWTVTSESLIQVFPEYGTVVGARVLYDGETGRSRG-YGFVCYSKRSELETALIS 196
Query: 261 TEKYEIDGKNLECSLAK 277
E++G+ + SLA+
Sbjct: 197 LNNVELEGRAIRVSLAE 247
>AV411398
Length = 427
Score = 46.2 bits (108), Expect = 9e-06
Identities = 27/101 (26%), Positives = 50/101 (48%)
Frame = +3
Query: 205 KNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKY 264
+N+P T + ++ LFE +G + +V L + + FV S A++AL E Y
Sbjct: 93 QNVPWTSTPEDVRSLFERYGTVLEVELSMYNKTRSRG-LAFVEMSSPEEALEALNKLESY 269
Query: 265 EIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGY 305
E +G+ L+ + A+P+ +++ +KPV + L Y
Sbjct: 270 EFEGRVLKLNYARPK-KKKAPPPVVQRKPVTFNLFVANLSY 389
Score = 39.7 bits (91), Expect = 8e-04
Identities = 29/111 (26%), Positives = 55/111 (49%), Gaps = 19/111 (17%)
Frame = +3
Query: 33 IANNV----SEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEELN 88
+A NV + ED+R + G V EV + ++ R G AFV + E A +A+ +L
Sbjct: 87 VAQNVPWTSTPEDVRSLFERYGTVLEVELSMYNKTRSR-GLAFVEMSSPEEALEALNKLE 263
Query: 89 NSEFKGKKIKCSSSQAKHR---------------LFIGNIPKKWTVEDMKK 124
+ EF+G+ +K + ++ K + LF+ N+ + T +D+++
Sbjct: 264 SYEFEGRVLKLNYARPKKKKAPPPVVQRKPVTFNLFVANLSYEATSKDLRE 416
>TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor SF2-like
protein, partial (46%)
Length = 597
Score = 45.8 bits (107), Expect = 1e-05
Identities = 34/121 (28%), Positives = 49/121 (40%)
Frame = +1
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
+ +YV NLP +I + ++ +F +G+I + L K + FV F + A A++
Sbjct: 256 RTIYVGNLPGDIRESEIEDIFYKYGRILDIEL---KVPPRPPCYCFVEFDNARDAEDAIR 426
Query: 260 NTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGY 319
+ Y DG L LA PT R GYG GG GG G
Sbjct: 427 GRDGYNFDGCRLRVELAHGGRG---------------PTSSDRRGYGGGGGGGGGGGGRI 561
Query: 320 G 320
G
Sbjct: 562 G 564
>TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG
polyadenylate-binding protein {Saccharomyces
cerevisiae;} , partial (18%)
Length = 518
Score = 45.4 bits (106), Expect = 1e-05
Identities = 22/59 (37%), Positives = 37/59 (62%)
Frame = +3
Query: 26 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
S +Y+ I +NVS+E+LR + G ++ ++MK E G +G+ FV F T E A++A+
Sbjct: 153 SNIYVKNIDDNVSDEELRGHFSACGSITYAKVMK-DEKGISQGFGFVCFSTPEEANKAV 326
Score = 35.4 bits (80), Expect = 0.015
Identities = 20/57 (35%), Positives = 31/57 (54%)
Frame = +3
Query: 202 VYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKAL 258
+YVKN+ +N++ + L+ F G IT + + G + FGFV FS A KA+
Sbjct: 159 IYVKNIDDNVSDEELRGHFSACGSITYAKVMKDEKGISQG-FGFVCFSTPEEANKAV 326
>AV409413
Length = 422
Score = 45.1 bits (105), Expect = 2e-05
Identities = 20/85 (23%), Positives = 45/85 (52%)
Frame = +2
Query: 30 IGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEELNN 89
+GGI +VSE++L+ F G+V E I++ + +G+ F+ F +++ + + N
Sbjct: 2 VGGIPTSVSEDELKGFFSKYGKVVEHEIIRDHTTKRSRGFGFIVFDNEKVVDTILTDGNM 181
Query: 90 SEFKGKKIKCSSSQAKHRLFIGNIP 114
+ G +++ ++ K G++P
Sbjct: 182 IDMAGTQVEIKKAEPKKSSNSGSLP 256
Score = 44.7 bits (104), Expect = 3e-05
Identities = 31/124 (25%), Positives = 53/124 (42%), Gaps = 3/124 (2%)
Frame = +2
Query: 204 VKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEK 263
V +P ++++D LK F +GK+ + + + + FGF+ F + L +
Sbjct: 2 VGGIPTSVSEDELKGFFSKYGKVVEHEIIRDHTTKRSRGFGFIVFDNEKVVDTILTDGNM 181
Query: 264 YEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVGGA--YGGIGA-GYG 320
++ G +E A+P + S S S P + P G G A YG G+ GYG
Sbjct: 182 IDMAGTQVEIKKAEP----KKSSNSGSLPPFASDSRPRSYNDGFSGFADSYGSFGSGGYG 349
Query: 321 AAGF 324
+
Sbjct: 350 PGSY 361
>TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(22%)
Length = 761
Score = 45.1 bits (105), Expect = 2e-05
Identities = 33/147 (22%), Positives = 65/147 (43%), Gaps = 8/147 (5%)
Frame = +1
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
++ +Y+G + NV++ L VG+V VR+ + + GY +V F A++A+
Sbjct: 328 TTSLYVGDLDQNVNDSQLYDLFNQVGQVVSVRVCRDLTTRRSLGYGYVNFSNPPDAARAL 507
Query: 85 EELNNSEFKGKKIKCSSS--------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISI 136
+ LN + + I+ S +FI N+ K + + + G ++S
Sbjct: 508 DVLNFTPLNNRPIRVMYSHRDPSIRKSGTANIFIKNLDKAIDHKALHDTFSSFG-SILSC 684
Query: 137 ELLKDLQSSGRNRGFAFIEYYNHACAE 163
++ D +SG+++ A CAE
Sbjct: 685 KIATD--ASGQSKAMALFSLTLSICAE 759
Score = 37.0 bits (84), Expect = 0.005
Identities = 32/134 (23%), Positives = 60/134 (43%)
Frame = +1
Query: 108 LFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQ 167
L++G++ + + + VG V+S+ + +DL ++ R+ G+ ++ + N A +
Sbjct: 337 LYVGDLDQNVNDSQLYDLFNQVGQ-VVSVRVCRDL-TTRRSLGYGYVNFSNPPDAARALD 510
Query: 168 KMSNSNFKLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKIT 227
+ NF NN P R+ S S +++KNL + I +L F G I
Sbjct: 511 VL---NFTPLNNRPIRVMYSHRDP-SIRKSGTANIFIKNLDKAIDHKALHDTFSSFGSIL 678
Query: 228 KVVLPPAKSGQEKS 241
+ SGQ K+
Sbjct: 679 SCKIATDASGQSKA 720
>TC12531 similar to UP|Q9ASQ8 (Q9ASQ8) At1g22910/F19G10_13, partial (39%)
Length = 688
Score = 44.3 bits (103), Expect = 3e-05
Identities = 18/58 (31%), Positives = 35/58 (60%)
Frame = -1
Query: 26 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQA 83
++V++GG+A +E ++ + + G++ E ++ K +G KGY FV F+ E A +A
Sbjct: 361 TKVFVGGLAWETQKETMKKYFEQFGDILEAVVITDKATGRSKGYGFVTFREPEAAMRA 188
>TC8015 similar to UP|ROC4_NICSY (P19683) 31 kDa ribonucleoprotein,
chloroplast precursor, partial (28%)
Length = 717
Score = 43.9 bits (102), Expect = 4e-05
Identities = 23/85 (27%), Positives = 41/85 (48%)
Frame = +3
Query: 201 AVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKN 260
+VYV NLP ++ L+++F HG + + + FGFV S + A+
Sbjct: 81 SVYVGNLPWSVDTARLEEIFREHGNVENARIVMDRETGRSRGFGFVTMSSEADINGAIAA 260
Query: 261 TEKYEIDGKNLECSLAKPEADQRSS 285
+ +DG+ + S+A+ + RSS
Sbjct: 261 LDGQSLDGRTIRVSVAEGRSGGRSS 335
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,450,665
Number of Sequences: 28460
Number of extensions: 87213
Number of successful extensions: 849
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 785
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 833
length of query: 370
length of database: 4,897,600
effective HSP length: 92
effective length of query: 278
effective length of database: 2,279,280
effective search space: 633639840
effective search space used: 633639840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0010.2


