

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
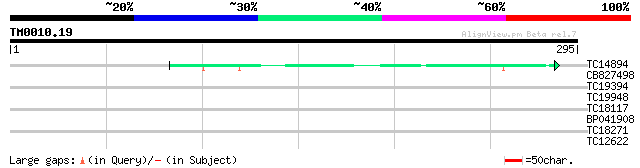
Query= TM0010.19
(295 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14894 weakly similar to UP|Q9LD82 (Q9LD82) ESTs AU029648, part... 49 1e-06
CB827498 28 1.4
TC19394 similar to UP|NSR1_YEAST (P27476) Nuclear localization s... 28 1.8
TC19948 weakly similar to UP|Q8DLX6 (Q8DLX6) Tlr0350 protein, pa... 27 3.1
TC18117 weakly similar to UP|Q9AYM8 (Q9AYM8) CPRD2 protein, part... 27 5.4
BP041908 26 7.0
TC18271 similar to UP|O23442 (O23442) Dynein light chain like pr... 26 9.1
TC12622 weakly similar to UP|AAQ62408 (AAQ62408) At3g43740, part... 26 9.1
>TC14894 weakly similar to UP|Q9LD82 (Q9LD82) ESTs AU029648, partial (84%)
Length = 982
Score = 48.9 bits (115), Expect = 1e-06
Identities = 58/224 (25%), Positives = 87/224 (37%), Gaps = 21/224 (9%)
Frame = +3
Query: 84 RLESRTLEEALMAVPD---LETVKFKVLSRRDNYEIRE-----------VEPYFVAEATM 129
R S TL+ A P +E + V+ + YEIR ++ + EAT
Sbjct: 90 RSGSLTLKNAGSIPPTCKRIECPSYDVIQVGNGYEIRRYNSTVWISNSPIQDISLVEATR 269
Query: 130 PGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAK 189
G F L Y+ GKN +K+EMT PV + + SDG
Sbjct: 270 TG------------FLRLFNYIQGKNDYSQKIEMTAPVLSEVSPSDGPFC---------- 383
Query: 190 MEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKL 249
+ + +SF +P AN P K + ++ VAV F GFV+D V + L
Sbjct: 384 ---ESSFVVSFFVPKVNQANPPPAK--GLHVQRWKPVNVAVRQFGGFVSDASVGEEAAAL 548
Query: 250 REALKS-------DGQFKIKKGTSVEVAQYNPPFTLPFQRRNEI 286
+ ++ + + + VAQY+ PF R NEI
Sbjct: 549 KASIAGTKWAAAIEKSHRAGHASVYSVAQYHAPFEYD-NRVNEI 677
>CB827498
Length = 558
Score = 28.5 bits (62), Expect = 1.4
Identities = 21/79 (26%), Positives = 33/79 (41%), Gaps = 7/79 (8%)
Frame = +2
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVA-------EATMPGKSGFDFSGASQSFNVLAEYLF 152
L+ + F +L R + I E F E + GK F + +FN LA LF
Sbjct: 143 LKRLMFFLLKSRREHVIPEFHIAFTELFESLEKELSKNGKVSFGEANDQAAFNFLARSLF 322
Query: 153 GKNTLKEKMEMTTPVYTSK 171
G N + ++++ P K
Sbjct: 323 GANPAETELDLDGPKLIQK 379
>TC19394 similar to UP|NSR1_YEAST (P27476) Nuclear localization sequence
binding protein (P67), partial (6%)
Length = 590
Score = 28.1 bits (61), Expect = 1.8
Identities = 19/69 (27%), Positives = 33/69 (47%)
Frame = -1
Query: 3 PLCNASLSSQSSIRTSIPGRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQAN 62
P C++S SS SS I+++ SSS ++ R S+ + SSLI ++ ++
Sbjct: 293 PPCSSSSSSASSAAAGSSSESG-GISSATSSSPELAGRATASSSSSSSSSLICSICCSSS 117
Query: 63 SLTQRLVVD 71
+ L D
Sbjct: 116 EELELLESD 90
>TC19948 weakly similar to UP|Q8DLX6 (Q8DLX6) Tlr0350 protein, partial (6%)
Length = 612
Score = 27.3 bits (59), Expect = 3.1
Identities = 14/51 (27%), Positives = 24/51 (46%)
Frame = -1
Query: 32 SSSERISNRRRTMSAVEARSSLILALASQANSLTQRLVVDVATETARYLFP 82
++ R+ RRR S S+I++L S ++ +TARY +P
Sbjct: 387 ANRSRVRRRRRARSPSSMAQSIIISLQC*PRSPCLNHTTELGIQTARYSYP 235
>TC18117 weakly similar to UP|Q9AYM8 (Q9AYM8) CPRD2 protein, partial (7%)
Length = 445
Score = 26.6 bits (57), Expect = 5.4
Identities = 14/31 (45%), Positives = 19/31 (61%)
Frame = -1
Query: 130 PGKSGFDFSGASQSFNVLAEYLFGKNTLKEK 160
PGKS F SG+SQ N+ + +TLK+K
Sbjct: 109 PGKS*FHASGSSQYPNLYNSHKLPLDTLKDK 17
>BP041908
Length = 487
Score = 26.2 bits (56), Expect = 7.0
Identities = 13/30 (43%), Positives = 19/30 (63%)
Frame = +3
Query: 10 SSQSSIRTSIPGRPNIAITNSASSSERISN 39
S+ ++ TS PG P A TNSA+S + S+
Sbjct: 240 STPTTTSTSPPGEPPPASTNSATSKKETSS 329
>TC18271 similar to UP|O23442 (O23442) Dynein light chain like protein,
partial (80%)
Length = 764
Score = 25.8 bits (55), Expect = 9.1
Identities = 20/59 (33%), Positives = 29/59 (48%), Gaps = 3/59 (5%)
Frame = +2
Query: 8 SLSSQSSI--RTSIP-GRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQANS 63
S SS SS+ ++SIP PN IT + + + RRRT S + R + S +S
Sbjct: 110 SSSSSSSVIFQSSIPFTNPNAIITLLEDTEK*VKTRRRTFSELLHRRGTTTPMTSNPHS 286
>TC12622 weakly similar to UP|AAQ62408 (AAQ62408) At3g43740, partial (25%)
Length = 668
Score = 25.8 bits (55), Expect = 9.1
Identities = 9/25 (36%), Positives = 15/25 (60%)
Frame = -2
Query: 144 FNVLAEYLFGKNTLKEKMEMTTPVY 168
F+ ++ Y F T K K+E T P++
Sbjct: 421 FSPISHYFFYNKTSKHKLEFTLPIF 347
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.129 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,601,022
Number of Sequences: 28460
Number of extensions: 36546
Number of successful extensions: 182
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 181
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 181
length of query: 295
length of database: 4,897,600
effective HSP length: 90
effective length of query: 205
effective length of database: 2,336,200
effective search space: 478921000
effective search space used: 478921000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0010.19


