

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.15
(268 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
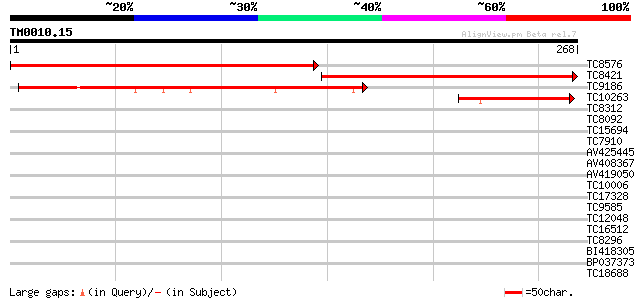
Score E
Sequences producing significant alignments: (bits) Value
TC8576 similar to UP|Q9SJN3 (Q9SJN3) At2g36220 protein (At2g3622... 302 3e-83
TC8421 similar to UP|Q9SJN3 (Q9SJN3) At2g36220 protein (At2g3622... 235 7e-63
TC9186 homologue to UP|Q9BIW6 (Q9BIW6) Brahma-associated protein... 185 6e-48
TC10263 67 3e-12
TC8312 similar to UP|Q8S901 (Q8S901) Syringolide-induced protein... 37 0.003
TC8092 similar to UP|Q8MLI6 (Q8MLI6) CG30127-PA, partial (3%) 33 0.066
TC15694 weakly similar to UP|Q8W042 (Q8W042) Prenylated Rab rece... 32 0.11
TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%) 31 0.25
AV425445 30 0.33
AV408367 30 0.33
AV419050 30 0.43
TC10006 similar to UP|P93396 (P93396) Transformer-SR ribonucleop... 30 0.56
TC17328 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol 4-r... 29 0.73
TC9585 similar to UP|O24078 (O24078) Protein phosphatase 2C, par... 29 0.73
TC12048 weakly similar to UP|Q9LGA3 (Q9LGA3) ESTs D22655(C0749),... 29 0.95
TC16512 similar to UP|Q9FXL4 (Q9FXL4) Elicitor inducible chitina... 29 0.95
TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1... 28 1.2
BI418305 28 2.1
BP037373 27 4.7
TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%) 27 4.7
>TC8576 similar to UP|Q9SJN3 (Q9SJN3) At2g36220 protein
(At2g36220/F2H17.17), partial (17%)
Length = 604
Score = 302 bits (774), Expect = 3e-83
Identities = 146/146 (100%), Positives = 146/146 (100%)
Frame = +1
Query: 1 MQQAETLVSACAGGSIDRKITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNALY 60
MQQAETLVSACAGGSIDRKITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNALY
Sbjct: 166 MQQAETLVSACAGGSIDRKITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNALY 345
Query: 61 ERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKR 120
ERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKR
Sbjct: 346 ERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKR 525
Query: 121 SVSVTKSVTSHAEPSSPKVSCMGRVR 146
SVSVTKSVTSHAEPSSPKVSCMGRVR
Sbjct: 526 SVSVTKSVTSHAEPSSPKVSCMGRVR 603
>TC8421 similar to UP|Q9SJN3 (Q9SJN3) At2g36220 protein
(At2g36220/F2H17.17), partial (10%)
Length = 749
Score = 235 bits (599), Expect = 7e-63
Identities = 120/121 (99%), Positives = 120/121 (99%)
Frame = +1
Query: 148 KRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSAA 207
KRDRSRSRSLRLRNSR SSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSAA
Sbjct: 1 KRDRSRSRSLRLRNSR*SSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRETDSAA 180
Query: 208 EDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGGMMRFASGRRSESWGVGESEIHVS 267
EDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGGMMRFASGRRSESWGVGESEIHVS
Sbjct: 181 EDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGGMMRFASGRRSESWGVGESEIHVS 360
Query: 268 R 268
R
Sbjct: 361 R 363
>TC9186 homologue to UP|Q9BIW6 (Q9BIW6) Brahma-associated protein 111kD,
partial (3%)
Length = 618
Score = 185 bits (470), Expect = 6e-48
Identities = 107/178 (60%), Positives = 126/178 (70%), Gaps = 13/178 (7%)
Frame = +1
Query: 5 ETLVSACAGGSIDRKITCETLANKTDPPERHPDSPPESFWLSKDEEHDWFDRNA-LYERK 63
ETLVSACAGGS DRKI CE L ++ +PP H DSPP SFWLS D E+DW+DRNA +YER
Sbjct: 43 ETLVSACAGGSTDRKIACEPLTDEPEPP-HHIDSPPASFWLSGDAEYDWWDRNAVVYERN 219
Query: 64 ESTKGSTS------SSTTNSQRFSLNL----KSRIIGLPKPQKPSFTDAKNRRNHKPCNN 113
EST+G++S S +NSQRFS NL K+ IIGLPK QK SF D +NRRNH N
Sbjct: 220 ESTRGNSSITSTNPGSNSNSQRFSKNLKHHPKAAIIGLPKSQKASFNDVRNRRNHHHPCN 399
Query: 114 IKLFPKRSVSV-TKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRN-SRKSSIDA 169
+LFPKRS SV +KS T EP SPKVSC+GRVRSKRDR+R R R+ S +S+ A
Sbjct: 400 ARLFPKRSASVRSKSDTVVIEPLSPKVSCIGRVRSKRDRNRRLRTRQRSISSTTSVSA 573
>TC10263
Length = 559
Score = 67.0 bits (162), Expect = 3e-12
Identities = 36/56 (64%), Positives = 40/56 (71%), Gaps = 1/56 (1%)
Frame = +2
Query: 213 TNSVVKARD-STASVNDASFAESISSEPPGLGGMMRFASGRRSESWGVGESEIHVS 267
T + KAR ST S D SF ES+ +EP GLG M RFASGRRSESWGVGE EI +S
Sbjct: 23 TRNSRKARGGSTVSRTDTSFEESVRTEPAGLGSMNRFASGRRSESWGVGECEIQLS 190
>TC8312 similar to UP|Q8S901 (Q8S901) Syringolide-induced protein 14-1-1,
partial (46%)
Length = 1358
Score = 37.4 bits (85), Expect = 0.003
Identities = 38/148 (25%), Positives = 58/148 (38%), Gaps = 14/148 (9%)
Frame = +2
Query: 133 EPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFR 192
EP+SPK+SCMG+++ K+++ + ++ K S+ E RK F
Sbjct: 401 EPTSPKISCMGQIKHKKNQIK------KSKSKKSVSLQNET-----RK----------FA 517
Query: 193 SGRRKKAHRETDSAAE---------DSTVTNSVVKARDSTASVNDASFAESISSEPPGLG 243
S K T ++ + S + A S S A S E P +
Sbjct: 518 STTNKTTTTSTSNSTDVIEVKKKNVASKFQRMLFHAAKQPKSAGRKSDASSAEREAPHMS 697
Query: 244 GMMRFASGR---RSESW--GVGESEIHV 266
M RF+SGR S W + EIH+
Sbjct: 698 QMKRFSSGRDAFSSFDWKAEMAAEEIHI 781
>TC8092 similar to UP|Q8MLI6 (Q8MLI6) CG30127-PA, partial (3%)
Length = 1073
Score = 32.7 bits (73), Expect = 0.066
Identities = 25/61 (40%), Positives = 31/61 (49%), Gaps = 3/61 (4%)
Frame = +2
Query: 106 RNHKPCNNIKLFPKRSVSVTKSVTSH-AEPSSPK--VSCMGRVRSKRDRSRSRSLRLRNS 162
R HKPC++ L RS S+T+S T H PSSP C RV ++ SL RN
Sbjct: 11 RAHKPCSSF*LAATRS-SLTESPTHHNG*PSSPPHLTPCFRRVTTRSTSLGETSLAYRNL 187
Query: 163 R 163
R
Sbjct: 188 R 190
>TC15694 weakly similar to UP|Q8W042 (Q8W042) Prenylated Rab receptor 2,
partial (46%)
Length = 458
Score = 32.0 bits (71), Expect = 0.11
Identities = 26/85 (30%), Positives = 37/85 (42%), Gaps = 4/85 (4%)
Frame = +3
Query: 86 SRIIGLPKPQKPSFTDAKNRRN--HKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMG 143
S + P K S T A+ R + PC+ + PKR++ + + +SP
Sbjct: 198 SSLASPPPSAKASPTAARGRSSLTAPPCHGLTPSPKRTLGSARISHTSVSTTSPSSPSRS 377
Query: 144 RVRSKRDRSR--SRSLRLRNSRKSS 166
R RS R RSR S S R R S+
Sbjct: 378 RCRSLRIRSRCLSSSASSRRGRSST 452
>TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%)
Length = 1035
Score = 30.8 bits (68), Expect = 0.25
Identities = 24/67 (35%), Positives = 35/67 (51%), Gaps = 3/67 (4%)
Frame = +2
Query: 110 PCNNIKLFPKRSVSVTKSVT--SHAEPSSPKVSCMGRV-RSKRDRSRSRSLRLRNSRKSS 166
P +L +R+V K+ SH +PS P+ GRV RS +RSR R RN++ S
Sbjct: 149 PGTEPQLHGRRTVGEEKAFQAFSHGQPSQPRFLHGGRVSRSLPHHARSRQHR-RNTQAHS 325
Query: 167 IDAGEEN 173
+ +G N
Sbjct: 326 VSSGTGN 346
>AV425445
Length = 344
Score = 30.4 bits (67), Expect = 0.33
Identities = 17/41 (41%), Positives = 22/41 (53%)
Frame = -2
Query: 70 TSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKP 110
T + + NS +SLNLK +P P K NR+NHKP
Sbjct: 124 TQN*SHNSSLYSLNLKK----IPFPWKTPSKHTLNRKNHKP 14
>AV408367
Length = 431
Score = 30.4 bits (67), Expect = 0.33
Identities = 17/59 (28%), Positives = 32/59 (53%)
Frame = +3
Query: 108 HKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDRSRSRSLRLRNSRKSS 166
H PC + KL PK++VS ++S + SP+ + ++ DR++S L+ S ++
Sbjct: 162 HAPCGHDKLDPKQTVSKMAKLSSASPVKSPQ--SLPAMKPNLDRNKSSKPLLKGSSNAT 332
>AV419050
Length = 409
Score = 30.0 bits (66), Expect = 0.43
Identities = 33/124 (26%), Positives = 48/124 (38%)
Frame = +1
Query: 144 RVRSKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFRAIFRSGRRKKAHRET 203
R RSKR RSRSRS+ SR + + E R R + +K HR
Sbjct: 91 RTRSKRSRSRSRSIDHGRSRHARSRSPSERSHR--------RRHHRTPSPDQPRKRHRR- 243
Query: 204 DSAAEDSTVTNSVVKARDSTASVNDASFAESISSEPPGLGGMMRFASGRRSESWGVGESE 263
DS +D T VV + F + G+++ + E+ G GE E
Sbjct: 244 DSVEDDHKDTKKVV-----------SDFVD----------GIVKEQQQKHKENGGGGEGE 360
Query: 264 IHVS 267
+ V+
Sbjct: 361 VEVN 372
>TC10006 similar to UP|P93396 (P93396) Transformer-SR ribonucleoprotein
(Fragment), partial (30%)
Length = 453
Score = 29.6 bits (65), Expect = 0.56
Identities = 26/87 (29%), Positives = 37/87 (41%), Gaps = 10/87 (11%)
Frame = +3
Query: 94 PQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMG---------- 143
P S +D+ RRN + + P R+ S ++S + + SP+
Sbjct: 78 PDSTSMSDSPVRRNSRSPS-----PWRAQSRSRSRSPRPKSRSPRPKSRSPRAKSRSPRP 242
Query: 144 RVRSKRDRSRSRSLRLRNSRKSSIDAG 170
R RS R RSRSRSL + R S G
Sbjct: 243 RSRSPRPRSRSRSLEKQRPRSRSRSRG 323
>TC17328 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol
4-reductase-like (Cinnamoyl-CoA reductase-like protein),
partial (21%)
Length = 479
Score = 29.3 bits (64), Expect = 0.73
Identities = 44/172 (25%), Positives = 64/172 (36%), Gaps = 10/172 (5%)
Frame = -3
Query: 27 NKTDPPERHPDSP----PESFWLSKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSL 82
+++ PP+ HP SP P S + ST G+TS+ST + S
Sbjct: 474 DESPPPQHHPASP*TPTPPS------------------PPRSSTPGTTSASTRRTPHPST 349
Query: 83 NLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRS---VSVTKSVTSHAEPSSPKV 139
R P S K+RR + + + R V ++ + E +
Sbjct: 348 TTHQR--PFPPASWDSGPMPKSRR*APRASRRRAWQLRGR*VFPVQRAGSGPCEDLPGRR 175
Query: 140 SCMGRVRSKRDRS---RSRSLRLRNSRKSSIDAGEENPGRIGRKHGFFERFR 188
+C GR R +R S R R R R R+ G PG GR G + R R
Sbjct: 174 ACRGRPRGRRRSSCGWRGRRRRSRGCRRGRW*CGNGLPG--GR*RGGWGRRR 25
>TC9585 similar to UP|O24078 (O24078) Protein phosphatase 2C, partial (31%)
Length = 502
Score = 29.3 bits (64), Expect = 0.73
Identities = 14/34 (41%), Positives = 19/34 (55%)
Frame = -3
Query: 146 RSKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGR 179
R + RSRSRS R R R+ ++ GE R+ R
Sbjct: 257 RRRGSRSRSRSTRRRRGRRVGLEVGERERERLRR 156
>TC12048 weakly similar to UP|Q9LGA3 (Q9LGA3) ESTs D22655(C0749), partial
(20%)
Length = 697
Score = 28.9 bits (63), Expect = 0.95
Identities = 32/138 (23%), Positives = 54/138 (38%), Gaps = 16/138 (11%)
Frame = -3
Query: 22 CETLANKTDPPERHP--DSPPESFWLSKDEEHDWFDRNA-------------LYERKEST 66
CE A++ P+ P SP E+ LS+ ++H + +Y+R+ S
Sbjct: 665 CEKEAHQQS*PQETP*TTSPMETQKLSQQKKHVQNNPQTASSEYPADSRTKPIYQREMSP 486
Query: 67 KGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTK 126
SST + + P PS + R+H P ++ L P S+ +
Sbjct: 485 DQKHQSSTPPPPQSP*ASSASSPSPSSPPPPSRSQPHPHRHHNPHHHTHLEPAASI*QNR 306
Query: 127 -SVTSHAEPSSPKVSCMG 143
+ H PS+P+ C G
Sbjct: 305 HQIQQHRPPSAPQ-QCAG 255
>TC16512 similar to UP|Q9FXL4 (Q9FXL4) Elicitor inducible chitinase
Nt-SubE76, partial (48%)
Length = 857
Score = 28.9 bits (63), Expect = 0.95
Identities = 33/140 (23%), Positives = 58/140 (40%), Gaps = 5/140 (3%)
Frame = +3
Query: 65 STKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAK-NRRNHKPC--NNIKLFPKRS 121
STK +++++T + RF L+S + P P P +D PC +++KL P
Sbjct: 45 STKLLSNTTSTMTSRFPPPLRSPLSETPTPPHPGRSDPNWYSPFSSPCSSSSVKLGPISW 224
Query: 122 VSVTKSVTSHAEPSSPKVSCM--GRVRSKRDRSRSRSLRLRNSRKSSIDAGEENPGRIGR 179
+ T S + P S + + GR+R + R ++RKS+ + P
Sbjct: 225 *TSTPSSRTSPTPMSSLSTTLFSGRIRVLQIRVTGCGTSTSSTRKSTPSSPRSPP*NSTT 404
Query: 180 KHGFFERFRAIFRSGRRKKA 199
+F R + R + A
Sbjct: 405 STSWFRRQAGLQRETKTSSA 464
>TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;}, partial (55%)
Length = 1276
Score = 28.5 bits (62), Expect = 1.2
Identities = 28/129 (21%), Positives = 47/129 (35%), Gaps = 6/129 (4%)
Frame = +2
Query: 46 SKDEEHDWFDRNALYERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNR 105
S+ +D ++R + ++ S S + S + + R P + D+++
Sbjct: 509 SRSRSYDKYERGDRHRGRDRDYRRRSRSRSASLDYKGRGRGRYDDEPSRSRSRSVDSRSP 688
Query: 106 RNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGR------VRSKRDRSRSRSLRL 159
P PKRSVS +S + P R RS R R + R
Sbjct: 689 ARRSPSPRRSPSPKRSVSPKRSTSPRKSPRGESPDNRSRDGRSPTPRSVSPRGRPVASRS 868
Query: 160 RNSRKSSID 168
+ RKS+ D
Sbjct: 869 PSPRKSNGD 895
>BI418305
Length = 437
Score = 27.7 bits (60), Expect = 2.1
Identities = 24/83 (28%), Positives = 36/83 (42%), Gaps = 1/83 (1%)
Frame = +1
Query: 55 DRNALYERKESTKGSTSSSTTNSQRFSLNLKSRIIGLPKPQKPSFTDAKNRRNHKPCNNI 114
D+ ++ G+T +ST N SL L T+A N +KP +
Sbjct: 76 DQRLTTRPNQAPHGATLTSTRNHGTLSLTL---------------TNAANGSPNKPTPTV 210
Query: 115 KLFPKRS-VSVTKSVTSHAEPSS 136
PKRS VS+ +S + A+P S
Sbjct: 211 NAAPKRSLVSMLRSKSFIAKPIS 279
>BP037373
Length = 488
Score = 26.6 bits (57), Expect = 4.7
Identities = 17/76 (22%), Positives = 32/76 (41%)
Frame = -3
Query: 92 PKPQKPSFTDAKNRRNHKPCNNIKLFPKRSVSVTKSVTSHAEPSSPKVSCMGRVRSKRDR 151
P P +P+FT + R +PC + F + ++P P ++
Sbjct: 282 PSPNRPNFTLSPPCRVRRPCPTLAPF------------TSSDPEGPSARNPSPPPTEPAA 139
Query: 152 SRSRSLRLRNSRKSSI 167
+S SL + N+ K+S+
Sbjct: 138 VKSASLAISNATKTSV 91
>TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%)
Length = 514
Score = 26.6 bits (57), Expect = 4.7
Identities = 32/132 (24%), Positives = 44/132 (33%), Gaps = 36/132 (27%)
Frame = +3
Query: 64 ESTKGSTSSSTTNSQRFSLNL-----------------KSRIIGLPKPQKPSFTDAKNRR 106
ES S+SSS+++S SLN + R G P P P+ A
Sbjct: 60 ESLSSSSSSSSSSSSPLSLNQISPPNAPPS*LSAPPSPEERFSGTPPPPPPATGSASTAT 239
Query: 107 NHKPCNNIKLFPKR-------------------SVSVTKSVTSHAEPSSPKVSCMGRVRS 147
P ++ P SVSV+ +H+ P+SP S
Sbjct: 240 PTPPTSSRSASPPSPSPANSHMASSPPSPTSALSVSVSTPSPAHSPPTSPPAPRSATSTS 419
Query: 148 KRDRSRSRSLRL 159
R S S RL
Sbjct: 420 SRISSPVSSRRL 455
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.126 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,521,101
Number of Sequences: 28460
Number of extensions: 62352
Number of successful extensions: 437
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 429
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 430
length of query: 268
length of database: 4,897,600
effective HSP length: 89
effective length of query: 179
effective length of database: 2,364,660
effective search space: 423274140
effective search space used: 423274140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0010.15


