

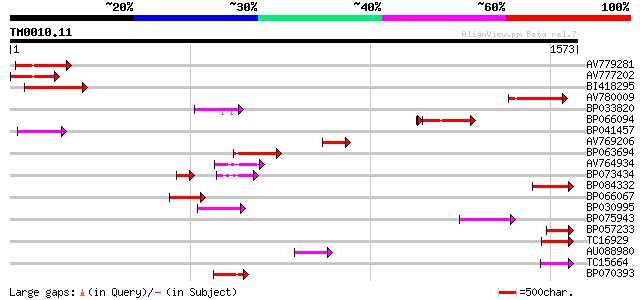
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.11
(1573 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV779281 235 4e-62
AV777202 218 5e-57
BI418295 214 9e-56
AV780009 152 5e-37
BP033820 117 1e-26
BP066094 112 1e-26
BP041457 112 5e-25
AV769206 97 3e-20
BP063694 96 6e-20
AV764934 91 1e-18
BP073434 91 1e-18
BP084332 89 7e-18
BP066067 88 9e-18
BP030995 88 9e-18
BP075943 86 3e-17
BP057233 86 5e-17
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 84 1e-16
AU088980 80 2e-15
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 78 1e-14
BP070393 76 5e-14
>AV779281
Length = 505
Score = 235 bits (600), Expect = 4e-62
Identities = 120/156 (76%), Positives = 133/156 (84%)
Frame = +1
Query: 15 PISVAIPQLSSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSD 74
PI + P LSS++LVH L +D+NFLSWKQHVEGIVRTHR QSFL H P++PPRFLSD
Sbjct: 46 PIPMVQPSLSSSALVHNAYL--EDTNFLSWKQHVEGIVRTHRWQSFLDH-PDVPPRFLSD 216
Query: 75 ADRVNSVENPAFLVWEQQDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSL 134
DRVN+VENPA+ +WEQQDSALFTWLLSSLSP+VLPTVVNC SWQ+ EAVLDFF AHS
Sbjct: 217 EDRVNAVENPAYQLWEQQDSALFTWLLSSLSPTVLPTVVNCSQSWQVREAVLDFFKAHSF 396
Query: 135 AQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALIS 170
AQSTQLRSELRSI KGSK SDYLKRIKSIVNA I+
Sbjct: 397 AQSTQLRSELRSIAKGSKTCSDYLKRIKSIVNASIT 504
>AV777202
Length = 421
Score = 218 bits (556), Expect = 5e-57
Identities = 106/138 (76%), Positives = 120/138 (86%)
Frame = +3
Query: 1 MAEEASPTQIPTPTPISVAIPQLSSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSF 60
MA+E + P P++V PQL+S+SLVHKL+LKLDDSNFLSWKQHVEGIVRTHRLQ+F
Sbjct: 9 MADEGHAQETAVPVPVAVLQPQLASSSLVHKLVLKLDDSNFLSWKQHVEGIVRTHRLQNF 188
Query: 61 LLHQPEIPPRFLSDADRVNSVENPAFLVWEQQDSALFTWLLSSLSPSVLPTVVNCQHSWQ 120
L+H P+IPPRFLS+ DRVN+VENPA+ VW+QQDSALFTWLLSSLS SVLPTVVNC SWQ
Sbjct: 189 LVH-PDIPPRFLSEEDRVNAVENPAYQVWDQQDSALFTWLLSSLSSSVLPTVVNCTQSWQ 365
Query: 121 IWEAVLDFFHAHSLAQST 138
IWEAVLDFF A SLAQST
Sbjct: 366 IWEAVLDFFQAQSLAQST 419
>BI418295
Length = 523
Score = 214 bits (545), Expect = 9e-56
Identities = 100/173 (57%), Positives = 136/173 (77%)
Frame = +2
Query: 42 LSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDADRVNSVENPAFLVWEQQDSALFTWLL 101
+ W+Q VEG +++H+L FL+ PEIP RFL++ DRV + ENPA+ W +QDS +FTWLL
Sbjct: 8 VGWRQQVEGAIQSHQLHDFLV-DPEIPQRFLTEEDRVAATENPAYTTWIKQDSMVFTWLL 184
Query: 102 SSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNTSDYLKRI 161
++LSP VLPTV+NC+ S Q+W +LDFF AQS +LRSEL++ITKGSK+ ++YL+RI
Sbjct: 185 TTLSPFVLPTVINCERSSQVWREILDFFENQCRAQSAKLRSELKNITKGSKSITEYLRRI 364
Query: 162 KSIVNALISIGDPVTYREHLEAIFDGLPEDYSHLMTIVTSREFPCSISQAEAM 214
K+IVN L+SIG+PV++R+HLEAIFDGLPEDYS L TI+ + +IS+ EAM
Sbjct: 365 KTIVNTLVSIGEPVSFRDHLEAIFDGLPEDYSALSTIIFTHASSSTISEVEAM 523
>AV780009
Length = 529
Score = 152 bits (383), Expect = 5e-37
Identities = 80/167 (47%), Positives = 107/167 (63%), Gaps = 2/167 (1%)
Frame = -1
Query: 1384 KAVKRILRYLKGTIHHGLHIRPCSLHSPVSLVAYCDADWGSDPDDRRSTSGSCVFFGPNL 1443
+A R+LRY+KG GL S SP+ L AY D+DW PD RRS +G +F G +L
Sbjct: 529 QAATRVLRYVKGAPAQGLFF---SADSPLKLQAYSDSDWAGCPDTRRSVTGYSIFLGTSL 359
Query: 1444 VTWTSKKQTLVARSSTEAEYRSLANTSAELLWIQSLLQ--ELQVPFTVPKVFCDNMGAVA 1501
++W +KKQT V+RSS+EAEYR+LA T E+ W+ L Q +L VP VP +FCDN A+
Sbjct: 358 ISWRTKKQTTVSRSSSEAEYRALAATVCEVQWLSYLFQFLKLNVPLPVP-LFCDNQSALH 182
Query: 1502 LTHNPVLHTRTKHMELDIFFVREKVQNQSLFVHHVPSVDQIADIFTK 1548
+ HNP H RTKH+ELD VR K+Q + + + + Q+ADIFTK
Sbjct: 181 IAHNPTFHERTKHIELDCHVVRAKLQAGLIHLLPISTHHQLADIFTK 41
>BP033820
Length = 537
Score = 117 bits (293), Expect = 1e-26
Identities = 68/160 (42%), Positives = 93/160 (57%), Gaps = 24/160 (15%)
Frame = -2
Query: 514 ASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTL 573
A+ S + + L+L LLLVP+ITKNLVSVS+FA+DN VYF FH C V QDT + L
Sbjct: 536 ATLPSLFNPKAILILNKLLLVPSITKNLVSVSRFAKDNNVYFTFHANFCAVRHQDTSELL 357
Query: 574 LRGSLGSDGLYSFDG----------MHSSNFHKDLPQVPVSSAFSLSKPA---------- 613
L GS+G DGLY FDG + +S F L P++ A S+P
Sbjct: 356 LTGSVGDDGLYYFDGPPSQAVSHSPVKASLFQSSLHSQPLAQAGLASQPVGNKASSAIDL 177
Query: 614 -SPAVNTIVSTS---NNVPSPTAYDLWHSRLGHPHYEALQ 649
S VN+ VS+S N+ + + Y+LWH+R+GHPH++ L+
Sbjct: 176 PSSLVNSSVSSSAPCNSSSNISIYELWHNRMGHPHHDVLK 57
>BP066094
Length = 532
Score = 112 bits (281), Expect(2) = 1e-26
Identities = 59/150 (39%), Positives = 94/150 (62%), Gaps = 3/150 (2%)
Frame = +2
Query: 1145 KPITIRLILTLAVSKRWHIHQLDVNNAFLHGALQEEVYMQQPAGFQ---NSDKTLVCKLN 1201
K IRL+++ +V+ +HQ++V +AFL+G + EEVY+ QP G + NSD + KL
Sbjct: 89 KTEAIRLLISFSVNHNIILHQMNVKSAFLNGYISEEVYVHQPPGXEDEKNSDH--IFKLK 262
Query: 1202 KALYGLKQAPRAWFDRLKAALVKYGFKASCCDPSLFTFHNKSNVIYILVYVDDIIITGNF 1261
K+LYGLKQAPRAW++RL + L++ D +LF K +++ + +YVDDII
Sbjct: 263 KSLYGLKQAPRAWYERLSSFLLENEXVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSAN 442
Query: 1262 LPFIQEVVQKLNSEFALKQLGQLDYFLGVQ 1291
+E + + +EF ++ +G+L YFLG+Q
Sbjct: 443 PSLCKEFSEMMQAEFEMRMMGELKYFLGIQ 532
Score = 25.4 bits (54), Expect(2) = 1e-26
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = +3
Query: 1128 YHQVKGFDYAETFSPVVK 1145
Y Q G DY ETF+PV +
Sbjct: 39 YSQQ*GIDYTETFAPVAR 92
>BP041457
Length = 495
Score = 112 bits (280), Expect = 5e-25
Identities = 55/136 (40%), Positives = 80/136 (58%)
Frame = +2
Query: 23 LSSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDADRVNSVE 82
L S + + KL FL WKQ VE I++ H L +++ +P IPPR+ SD + N V
Sbjct: 86 LRSALFSYLIFEKLSAKKFLLWKQQVEPIIKAHHLHCYIV-EPAIPPRYSSDEEHANGVV 262
Query: 83 NPAFLVWEQQDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRS 142
NPA+ +WE D L +WL S LS SV ++ C +S+Q+W + FFH + A+STQ+R+
Sbjct: 263 NPAYPIWETHDQFLLSWLQSLLSSSVHTCMIGCVNSFQLWVKIHSFFHNQTRAKSTQIRT 442
Query: 143 ELRSITKGSKNTSDYL 158
ELRS G + Y+
Sbjct: 443 ELRSAKLGGRLLHGYI 490
>AV769206
Length = 536
Score = 96.7 bits (239), Expect = 3e-20
Identities = 46/79 (58%), Positives = 58/79 (73%), Gaps = 2/79 (2%)
Frame = -3
Query: 868 LRPYNSSKLSFRSQECVFLGYSSSHKGYKCLASDGRIYISKDVQFNEFKFPYSTLFSSEF 927
LRPYNS+KLS S+ECVF+GYS +HKGYKCL GRIYISKDV F+E +FPY +LF E
Sbjct: 531 LRPYNSTKLSLHSKECVFIGYSINHKGYKCLDQSGRIYISKDVLFHEHRFPYPSLFPDET 352
Query: 928 PASNPSSV--SSVIPVISY 944
+S + S IP++S+
Sbjct: 351 TSSTSAEFFPLSTIPIVSH 295
>BP063694
Length = 511
Score = 95.5 bits (236), Expect = 6e-20
Identities = 53/135 (39%), Positives = 83/135 (61%), Gaps = 1/135 (0%)
Frame = -2
Query: 620 IVSTSNNVPSPTAYDLWHSRLGHPHYEALQSALTTCKIPVPH-KSKYTICHSCCVAKSHR 678
+++TS+ +P +++LWHSRLGH +++ ++ + V K C SC +AKS R
Sbjct: 402 LLTTSSPLPR-ASFELWHSRLGHVNFDIIKQLHKHGCLDVSSILPKPICCTSCQMAKSKR 226
Query: 679 LPSSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETST 738
L ++ +A L+LI DL GP+ + S G+SYF+ VD FSRFTW Y LKRKS+ S
Sbjct: 225 LVFHDNNKRASAVLDLIHCDLRGPSPVASIDGFSYFVIFVDDFSRFTWFYPLKRKSDFSD 46
Query: 739 VFIQFQAMVELKFGL 753
V ++F+ +E +F +
Sbjct: 45 VLLRFKVFMENRFSV 1
>AV764934
Length = 406
Score = 91.3 bits (225), Expect = 1e-18
Identities = 51/139 (36%), Positives = 73/139 (51%), Gaps = 2/139 (1%)
Frame = -2
Query: 569 TCKTLLRGSLGSDGLYSFD--GMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNN 626
T + L+ G +G DGLYSF + + + + PVS P+S + +ST
Sbjct: 372 TSEPLIHGVVGPDGLYSFPLPQLQQLGYTRSMSSSPVSGRH----PSSVSAFVTLST--- 214
Query: 627 VPSPTAYDLWHSRLGHPHYEALQSALTTCKIPVPHKSKYTICHSCCVAKSHRLPSSASST 686
WHSRLGHPH + L+ L++C + K +CC+ K+HRLPS S+T
Sbjct: 213 ---------WHSRLGHPHLDNLKRVLSSCNVSYSPKDTVEFRTACCLGKAHRLPSQMSTT 61
Query: 687 LYTAPLELIFADLWGPASI 705
Y P ELI++DLWGPA +
Sbjct: 60 TY-LPFELIYSDLWGPAPV 7
>BP073434
Length = 490
Score = 90.9 bits (224), Expect = 1e-18
Identities = 49/117 (41%), Positives = 71/117 (59%)
Frame = -1
Query: 573 LLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTA 632
LL G +G+DGLYSF G+ +S L V S +F + VNTI +T+ N
Sbjct: 310 LLLGHVGADGLYSFPGLLAS-----LSSVDRSPSFDI-------VNTITTTTTN-----K 182
Query: 633 YDLWHSRLGHPHYEALQSALTTCKIPVPHKSKYTICHSCCVAKSHRLPSSASSTLYT 689
+ +WHSRLGHPH +ALQ+ L +C I + + C++CC+ KSH LPSS S+++Y+
Sbjct: 181 FLIWHSRLGHPHQDALQAVLQSCNIHLSSRDMTDFCYACCLGKSHILPSSLSTSVYS 11
Score = 50.1 bits (118), Expect = 3e-06
Identities = 22/51 (43%), Positives = 31/51 (60%)
Frame = -2
Query: 462 APSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIG 512
A + +W+PDSGA+HHVT + I G D +++GNGQGL + S G
Sbjct: 477 AAGLVASWYPDSGASHHVTPHGNNIQYPDPFEGPDHIFVGNGQGLSVASSG 325
>BP084332
Length = 368
Score = 88.6 bits (218), Expect = 7e-18
Identities = 44/112 (39%), Positives = 70/112 (62%)
Frame = +2
Query: 1451 QTLVARSSTEAEYRSLANTSAELLWIQSLLQELQVPFTVPKVFCDNMGAVALTHNPVLHT 1510
Q+ +A S+ EAEY S A S ++LW++ L++ Q+ + ++CDN A++L+ NP+LH+
Sbjct: 32 QSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHS 211
Query: 1511 RTKHMELDIFFVREKVQNQSLFVHHVPSVDQIADIFTKALSPTRFEDLRSKL 1562
R KH+E+ F+R+ VQ L + V + Q ADIFTK L+ RF + L
Sbjct: 212 RAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNL 367
>BP066067
Length = 505
Score = 88.2 bits (217), Expect = 9e-18
Identities = 45/100 (45%), Positives = 65/100 (65%)
Frame = -2
Query: 443 LPPRAPAPQIRAPHAMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGN 502
LP +PAP + + +AP T +W DSGA+HHVT+D + + +++ G +QVY+ +
Sbjct: 369 LPQSSPAPMAFVSNTV--AAP*TTHSW*ADSGASHHVTNDPQHLMESITMPGQEQVYVCS 196
Query: 503 GQGLPIQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLV 542
GQGLPI SIG SFSSP+ + T LK+ +P ITKN+V
Sbjct: 195 GQGLPILSIGFTSFSSPLISNTKFSLKDFFHLPTITKNIV 76
>BP030995
Length = 458
Score = 88.2 bits (217), Expect = 9e-18
Identities = 54/132 (40%), Positives = 74/132 (55%), Gaps = 1/132 (0%)
Frame = -3
Query: 522 TQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSD 581
T L+L NLL VP + KNL SVS F + N V+FEFH C V SQ+T +TLL+G+L +
Sbjct: 438 TSFPLVLNNLLHVPGVCKNLSSVSXFCKQNNVFFEFHSDSCLVKSQETEETLLQGTL-RN 262
Query: 582 GLYSFDG-MHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWHSRL 640
GL +FD +H N H + S+ S S PAS ++ S++ + LWHSRL
Sbjct: 261 GLCAFDNLLHKVNLH-SASTLSNSALLSSSSPASALSVSVFSSNKCAQADIDSRLWHSRL 85
Query: 641 GHPHYEALQSAL 652
GH + + L
Sbjct: 84 GHASQKIVSHVL 49
>BP075943
Length = 547
Score = 86.3 bits (212), Expect = 3e-17
Identities = 52/155 (33%), Positives = 83/155 (53%), Gaps = 1/155 (0%)
Frame = -3
Query: 1248 ILVYVDDIIITGNFLPFIQEVVQKLNSEFALKQLGQLDYFLGVQVHHLQDRSLLLTQTKY 1307
+L+YVDD+++ GN + IQ V L+ +F +K LG+ YFLG+++ ++L Q KY
Sbjct: 521 LLLYVDDVLLAGNDMHEIQLVKSSLHDQFRIKDLGEAKYFLGLEIAR-STSGIVLNQRKY 345
Query: 1308 IGDLL-ERADMAEAKGISTPMVSGARLSKHGADYFSDPTLYRSIVGALQYATLTRPEISF 1366
L+ + + + L + +D YR IVG L Y TRP+I+F
Sbjct: 344 ALQLISDSGHFGFQPRFYSHGTNSQTLGTNTGTPLTDIGSYRRIVGRLLYLNTTRPDITF 165
Query: 1367 SVNKVCQFLSQPLEEHWKAVKRILRYLKGTIHHGL 1401
+VN++ QFLS P + H + + L+YL G+ GL
Sbjct: 164 AVNQLSQFLSAPTDIHEQQLTGFLKYL*GSPGSGL 60
>BP057233
Length = 473
Score = 85.9 bits (211), Expect = 5e-17
Identities = 39/75 (52%), Positives = 53/75 (70%)
Frame = -2
Query: 1490 PKVFCDNMGAVALTHNPVLHTRTKHMELDIFFVREKVQNQSLFVHHVPSVDQIADIFTKA 1549
P ++CDN+ A AL NPVLH R+KH+E+D+ ++R++V + V +VP+ DQIAD TK
Sbjct: 451 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNEVVVAYVPTTDQIADCLTKP 272
Query: 1550 LSPTRFEDLRSKLNV 1564
LS TRF LR KL V
Sbjct: 271 LSHTRFSQLRDKLGV 227
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 84.3 bits (207), Expect = 1e-16
Identities = 41/91 (45%), Positives = 59/91 (64%), Gaps = 1/91 (1%)
Frame = +1
Query: 1475 WIQSLLQELQVPFTVPK-VFCDNMGAVALTHNPVLHTRTKHMELDIFFVREKVQNQSLFV 1533
W+ LLQ+L+VPF P V+CDN A + NPV H RTKH+E+D VRE++Q + +
Sbjct: 4 WLTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDCHIVRERIQKGLIHL 183
Query: 1534 HHVPSVDQIADIFTKALSPTRFEDLRSKLNV 1564
+ S + +ADI+TKALSP F + +KL +
Sbjct: 184 LPISSSEPLADIYTKALSPQNFHQICAKLGL 276
>AU088980
Length = 360
Score = 80.5 bits (197), Expect = 2e-15
Identities = 41/105 (39%), Positives = 58/105 (55%)
Frame = +2
Query: 791 NGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKV 850
N VERKHRHI+ +L+ + +P + W A A +LIN LP+P+L PY L
Sbjct: 2 NSIVERKHRHILNVTRALMFHSYLPKNLWTFAVKHAAYLINXLPSPLLKGQCPYQFLNND 181
Query: 851 PADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGY 895
L+VFG+ CY +N KL R+++CVFLG+ + GY
Sbjct: 182 SPTLLDLKVFGTLCYSSTLTHNXQKLDPRARKCVFLGFKTGTXGY 316
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 78.2 bits (191), Expect = 1e-14
Identities = 42/94 (44%), Positives = 57/94 (59%), Gaps = 1/94 (1%)
Frame = +1
Query: 1472 ELLWIQSLLQELQVPFTVPKV-FCDNMGAVALTHNPVLHTRTKHMELDIFFVREKVQNQS 1530
EL W+ +LQ+L+VPF P + +CD+ A + N V H RTKH+++D VREK+Q +
Sbjct: 4 EL*WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDCHVVREKLQAKL 183
Query: 1531 LFVHHVPSVDQIADIFTKALSPTRFEDLRSKLNV 1564
+ + SVDQ ADI TK L F L SKL V
Sbjct: 184 FHLLPISSVDQTADILTKPLESGPFSHLVSKLGV 285
>BP070393
Length = 359
Score = 75.9 bits (185), Expect = 5e-14
Identities = 39/100 (39%), Positives = 61/100 (61%), Gaps = 4/100 (4%)
Frame = -3
Query: 566 SQDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVS----SAFSLSKPASPAVNTIV 621
+++T K LLRGS+GSDGLY+FD +++++ + +S S S P + +
Sbjct: 294 AKETSKVLLRGSVGSDGLYTFDDVNAASVSSSCSKSDLSTSTPSVLSTVYPGTGKSQNMS 115
Query: 622 STSNNVPSPTAYDLWHSRLGHPHYEALQSALTTCKIPVPH 661
S S+ S +YDLWH RLGHPH+E L++ L C +P+P+
Sbjct: 114 SVSSLGNS--SYDLWHCRLGHPHHEVLKTTLQLCNVPLPN 1
Score = 30.0 bits (66), Expect = 3.0
Identities = 13/23 (56%), Positives = 15/23 (64%)
Frame = -2
Query: 483 ASAISDGMSVTGNDQVYMGNGQG 505
A SD S TG +Q+ MGNGQG
Sbjct: 355 AGNFSDTSSFTGTEQILMGNGQG 287
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,305,340
Number of Sequences: 28460
Number of extensions: 548568
Number of successful extensions: 9493
Number of sequences better than 10.0: 583
Number of HSP's better than 10.0 without gapping: 5546
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7933
length of query: 1573
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1471
effective length of database: 1,994,680
effective search space: 2934174280
effective search space used: 2934174280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0010.11


