

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.10
(289 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
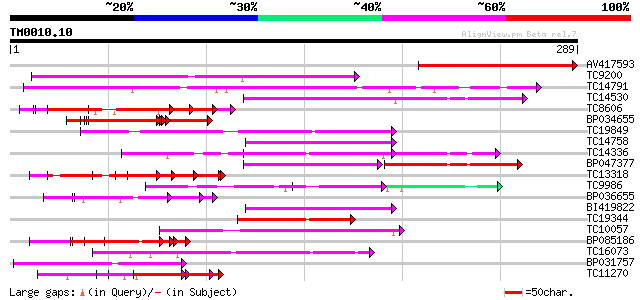
Score E
Sequences producing significant alignments: (bits) Value
AV417593 117 3e-27
TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, ... 80 4e-16
TC14791 similar to UP|Q9LJ04 (Q9LJ04) ESTs AU082210(C53655), par... 64 4e-11
TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribos... 60 4e-10
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 55 1e-08
BP034655 55 1e-08
TC19849 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, p... 54 3e-08
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 54 4e-08
TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 54 4e-08
BP047377 53 7e-08
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 52 2e-07
TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding prot... 50 3e-07
BP036655 50 6e-07
BI419822 50 6e-07
TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG polyaden... 48 2e-06
TC10057 similar to UP|Q41124 (Q41124) Chloroplast RNA binding pr... 48 2e-06
BP085186 48 2e-06
TC16073 similar to UP|Q9FJN9 (Q9FJN9) Poly(A)-binding protein II... 47 5e-06
BP031757 46 6e-06
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 46 6e-06
>AV417593
Length = 423
Score = 117 bits (292), Expect = 3e-27
Identities = 51/81 (62%), Positives = 67/81 (81%)
Frame = +2
Query: 209 WTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKL 268
W ED++K V + GPGV ++EL+KD+++ NRGFAFI+YYNHACAE+SRQKM + FKL
Sbjct: 5 WGEEDLRKTVTEAGPGVTAVELVKDVKNINNNRGFAFIDYYNHACAEHSRQKMMSPTFKL 184
Query: 269 ENNAPTVSWADPRNSESSAVS 289
+NAPTVSWADP+N++SSA S
Sbjct: 185 GDNAPTVSWADPKNADSSASS 247
>TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(31%)
Length = 727
Score = 80.1 bits (196), Expect = 4e-16
Identities = 54/185 (29%), Positives = 89/185 (47%), Gaps = 18/185 (9%)
Frame = +1
Query: 12 VTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEE 71
+T + P+ + E P +++ + +E E E EE EEEE E+EE EE EEEE
Sbjct: 103 ITKKRKLVPKSSQSAELPFKLQNKLEAEVQEYEEVEEEVEEEVEEEEEEEEEEEEGEEEE 282
Query: 72 EEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHG-------------- 117
EEEEEEEEVE E++ D E + + + K++ LLA
Sbjct: 283 EEEEEEEEVEAEAEEEDDEP---IQKLLEPMGKEQIISLLADAASNHRDVADRIRKVADE 453
Query: 118 ----SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
++++ G+ + + L Q G + + + + K +G+ KGY F+ FK++ A
Sbjct: 454 DVSHRKIFVHGLGWDTTATTLVYAFQQYGAIEDCKAVTDKVTGKSKGYGFILFKSRRGAR 633
Query: 174 QAIEE 178
A++E
Sbjct: 634 NALKE 648
>TC14791 similar to UP|Q9LJ04 (Q9LJ04) ESTs AU082210(C53655), partial (37%)
Length = 1779
Score = 63.5 bits (153), Expect = 4e-11
Identities = 67/317 (21%), Positives = 131/317 (41%), Gaps = 53/317 (16%)
Frame = +2
Query: 8 RQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVED------ 61
+ + A + E E + E E D+ V+ + D + E E +++E +ED
Sbjct: 197 QSNNTMAVDEEPEEQDQETEDQPQIDDAVEEEQDPEAPATTESEPQQQQETLEDNADAPN 376
Query: 62 -----------EEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEK--KKHA 108
E V EEEEEEE++E E +P E + T +++ K+ A
Sbjct: 377 ESSAAAAAAASESVANGSNNNEEEEEEEDLELEEEPV---ERLLEPFTREQLHSLVKQAA 547
Query: 109 E-----------LLALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGE 157
E L + P ++++ G+ + + E L GE+ + + + K +G+
Sbjct: 548 EKYPDFVENVRQLADVDPAHRKIFVHGLGWDTTAETLTSVFSKYGEIEDCKAVTDKVTGK 727
Query: 158 GKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCS------------------SSQAKHR 199
KGYAF+ FK ++ +A++ N + G + S S + +
Sbjct: 728 SKGYAFILFKHRDGCRRALK--NPQKKIGNRTTSSQLASAGPVPATPPNAPPVSEYTQRK 901
Query: 200 LFIGNIPKKWTVEDMK-----KVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACA 254
+F+ N+ +E +K K +V G + ++ +++GR +GFA Y +
Sbjct: 902 IFVSNVSS--DIEPLKLLEFFKQFGEVEDGPLGLD-----KNTGRPKGFALFVYRS---V 1051
Query: 255 EYSRQKMSNSNFKLENN 271
E +++ + N + E +
Sbjct: 1052ESAKKALEEPNKEYEGH 1102
>TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribosomal
protein 2, chloroplast precursor (PSRP-2), partial (68%)
Length = 1104
Score = 60.1 bits (144), Expect = 4e-10
Identities = 38/165 (23%), Positives = 82/165 (49%), Gaps = 20/165 (12%)
Frame = +2
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+Y+G I ++ ++L Q V + +M K SG + +AFV KT E A+ AIE+L
Sbjct: 236 LYVGNIPRTLTNDELANIVQEHAAVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKL 415
Query: 180 NNSEFKGKKIKCSSSQ--------------------AKHRLFIGNIPKKWTVEDMKKVVA 219
N ++ G++IK + ++ + +++++GN+ K T + +K +
Sbjct: 416 NGTQIGGREIKVNVTEKPLTTVDFPLVQAEESEYVDSPYKVYVGNLAKTVTSDMLKNFFS 595
Query: 220 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNS 264
+ G V+S ++ + + + ++ GF F+ + + E + +N+
Sbjct: 596 EKGK-VLSAKVSR-VPGTSKSSGFGFVTFSSDEDVEAAISSFNNA 724
Score = 37.0 bits (84), Expect = 0.004
Identities = 20/55 (36%), Positives = 35/55 (63%)
Frame = +2
Query: 192 SSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFI 246
+ S+A RL++GNIP+ T +++ +V + V E++ D + SGR+R FAF+
Sbjct: 212 TQSEADRRLYVGNIPRTLTNDELANIVQE-HAAVEKAEVMYD-KYSGRSRRFAFV 370
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 55.5 bits (132), Expect = 1e-08
Identities = 28/65 (43%), Positives = 42/65 (64%)
Frame = -2
Query: 20 PEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEE 79
PE N E E+ D ++Q D D D++ ++E E++ EDEE E VEEE+ E+EEE+E
Sbjct: 446 PEENGEDEEDEDGEDQEDDDDDDEDDDE-------EDDGGEDEEEEGVEEEDNEDEEEDE 288
Query: 80 VEEES 84
+EE+
Sbjct: 287 EDEEA 273
Score = 46.6 bits (109), Expect = 5e-06
Identities = 33/131 (25%), Positives = 59/131 (44%), Gaps = 28/131 (21%)
Frame = -2
Query: 13 TAQNPESPEGNSEPEKPTDSDEQVDFDGDN---DQEEEIEYEE----------------- 52
T E + ++E E D + D DGD + EE++ E+
Sbjct: 656 TKSQSEDKQHDAEGEDGNDDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKA 477
Query: 53 --------VEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEK 104
EE EDEE E+ E++E++++++E+ +EE + EEE V+ ++E E+
Sbjct: 476 PEGGPGAGAGPEENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEE 297
Query: 105 KKHAELLALPP 115
+ + AL P
Sbjct: 296 EDEEDEEALQP 264
Score = 44.7 bits (104), Expect = 2e-05
Identities = 25/81 (30%), Positives = 48/81 (58%)
Frame = -2
Query: 14 AQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEE 73
+ + ++PEG P +E + + D D E++ E+ ++++E +DEE + E+EEEE
Sbjct: 494 SNSKKAPEGG--PGAGAGPEENGEDEEDEDGEDQ---EDDDDDDEDDDEEDDGGEDEEEE 330
Query: 74 EEEEEEVEEESKPSDGEEEMR 94
EEE+ E+E + + EE ++
Sbjct: 329 GVEEEDNEDEEEDEEDEEALQ 267
Score = 42.7 bits (99), Expect = 7e-05
Identities = 23/66 (34%), Positives = 41/66 (61%)
Frame = -2
Query: 41 DNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTND 100
+N ++EE E E +E+++ +DE+ +E E++ E+EEEE VEEE + E+E +
Sbjct: 440 ENGEDEEDEDGEDQEDDDDDDEDDDE-EDDGGEDEEEEGVEEEDNEDEEEDEEDEEALQP 264
Query: 101 EVEKKK 106
++KK
Sbjct: 263 PKKRKK 246
Score = 39.3 bits (90), Expect = 8e-04
Identities = 26/103 (25%), Positives = 52/103 (50%), Gaps = 2/103 (1%)
Frame = -2
Query: 6 GTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVE 65
G + D+++++ NS + + + EE E EE E+ E+ ED++ +
Sbjct: 560 GEGEEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDEEDEDGEDQEDDDDD 381
Query: 66 EVEEEEEEE--EEEEEVEEESKPSDGEEEMRVDHTNDEVEKKK 106
+ +++EE++ E+EEE E + ++ EEE D + KK+
Sbjct: 380 DEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDEEALQPPKKR 252
>BP034655
Length = 517
Score = 55.1 bits (131), Expect = 1e-08
Identities = 31/64 (48%), Positives = 42/64 (65%), Gaps = 1/64 (1%)
Frame = +1
Query: 41 DNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDG-EEEMRVDHTN 99
D+D+EEE E EE EEEEE E+EE EE E+EE++EE+E+E PS + + T+
Sbjct: 136 DDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDEDYIPVAPSHRVSPNVHQNPTS 315
Query: 100 DEVE 103
D VE
Sbjct: 316 DSVE 327
Score = 53.9 bits (128), Expect = 3e-08
Identities = 26/43 (60%), Positives = 33/43 (76%)
Frame = +1
Query: 37 DFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEE 79
DF +D EEE E EE EEEEE E+EE EE EE+EE++EE+E+
Sbjct: 121 DFGLSDDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDED 249
Score = 53.5 bits (127), Expect = 4e-08
Identities = 26/43 (60%), Positives = 34/43 (78%)
Frame = +1
Query: 40 GDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEE 82
G +D +EE E EE EEEEE E+EE EE EEE+EE++EE+E E+
Sbjct: 127 GLSDDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Score = 52.4 bits (124), Expect = 9e-08
Identities = 24/42 (57%), Positives = 33/42 (78%)
Frame = +1
Query: 39 DGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEV 80
D D ++EEE E EE EEEEE E+EE EE EE++EE+E+E+ +
Sbjct: 136 DDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDEDYI 261
Score = 48.9 bits (115), Expect = 1e-06
Identities = 23/48 (47%), Positives = 35/48 (72%)
Frame = +1
Query: 30 TDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEE 77
T +D + D + ++EEE E EE EEEEE E+EE E+ E++EE+E+E+
Sbjct: 112 TAADFGLSDDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Score = 33.1 bits (74), Expect = 0.057
Identities = 13/41 (31%), Positives = 28/41 (67%)
Frame = +1
Query: 23 NSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEE 63
+ + E+ + +E+ + + + ++EEE E E+ E++EE EDE+
Sbjct: 133 SDDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Score = 32.0 bits (71), Expect = 0.13
Identities = 17/65 (26%), Positives = 32/65 (49%)
Frame = +1
Query: 72 EEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSE 131
+++EEEEE EEE + + EEE + DE + ++ + +P S + N +
Sbjct: 136 DDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDEDYIPVAPSHRVSPNVHQNPTS 315
Query: 132 EDLRV 136
+ + V
Sbjct: 316 DSVEV 330
Score = 32.0 bits (71), Expect = 0.13
Identities = 13/59 (22%), Positives = 34/59 (57%)
Frame = +1
Query: 9 QRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEV 67
+ ++ A + + + + + +E+ + + + ++EEE E EE +EE++ EDE+ + +
Sbjct: 85 RNEIAAAHLTAADFGLSDDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDEDYI 261
>TC19849 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, partial
(30%)
Length = 603
Score = 53.9 bits (128), Expect = 3e-08
Identities = 40/161 (24%), Positives = 73/161 (44%)
Frame = +2
Query: 37 DFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVD 96
D DG + + +E +E + D ++ +++E V + K S+ E E++
Sbjct: 29 DVDGKSKCFGFVNFENADEAAKAVD-----ALNGKKFDDKEWYVGKAKKKSERELELKEQ 193
Query: 97 HTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESG 156
H E + G+ +Y+ + +NV +E LR G ++ +IM+ G
Sbjct: 194 HEQSMQETVDKYQ-------GANLYLKNLDDNVDDEKLRELFSEFGTITSCKIMRDPH-G 349
Query: 157 EGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAK 197
+G FVAF T E A++A+ E+N GK + + +Q K
Sbjct: 350 VSRGSGFVAFSTPEEATRALGEMNGKMVDGKPLYVALAQKK 472
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 53.5 bits (127), Expect = 4e-08
Identities = 23/77 (29%), Positives = 43/77 (54%)
Frame = +1
Query: 121 YIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELN 180
++GG+A L S GE+ + +++ +E+G +G+ FV F ++E AIE +N
Sbjct: 91 FVGGLAWTTDSHTLEQAFSSYGEIIDSKVVNDRETGRSRGFGFVTFTSEEAMRSAIEGMN 270
Query: 181 NSEFKGKKIKCSSSQAK 197
+E G+ I + +QA+
Sbjct: 271 GNELDGRNITVNEAQAR 321
Score = 27.7 bits (60), Expect = 2.4
Identities = 20/90 (22%), Positives = 47/90 (52%), Gaps = 2/90 (2%)
Frame = +1
Query: 194 SQAKHRLFIGNIPKKWTVED--MKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNH 251
+ ++R F+G + WT + +++ + G +I +++ D + +GR+RGF F+ + +
Sbjct: 70 ADVEYRCFVGGLA--WTTDSHTLEQAFSSYGE-IIDSKVVND-RETGRSRGFGFVTFTSE 237
Query: 252 ACAEYSRQKMSNSNFKLENNAPTVSWADPR 281
+ + M+ + +L+ TV+ A R
Sbjct: 238 EAMRSAIEGMNGN--ELDGRNITVNEAQAR 321
>TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(85%)
Length = 1917
Score = 53.5 bits (127), Expect = 4e-08
Identities = 35/143 (24%), Positives = 71/143 (49%), Gaps = 12/143 (8%)
Frame = +2
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
++I + + + L + G + ++ SG+ KGY FV F+T+E A AIE+L
Sbjct: 98 IFIKNLDKAIDHKALHDTFSTFGNILSCKVA-TDSSGQSKGYGFVQFETEEAAQNAIEKL 274
Query: 180 NNSEFKGKKI-----------KCSSSQAK-HRLFIGNIPKKWTVEDMKKVVADVGPGVIS 227
N K++ + S+ +AK + +F+ N+ + T +++K V + G + S
Sbjct: 275 NGMLLNDKQVYVGPFLRKQERESSTDKAKFNNVFVKNLSESTTDDELKNVFGEFGT-ITS 451
Query: 228 IELLKDLQSSGRNRGFAFIEYYN 250
+++D G+++ F F+ + N
Sbjct: 452 AVVMRD--GDGKSKCFGFVNFEN 514
Score = 50.1 bits (118), Expect = 4e-07
Identities = 37/142 (26%), Positives = 72/142 (50%), Gaps = 2/142 (1%)
Frame = +2
Query: 58 EVEDEEVEEVEEEEEEEEEEEE--VEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPP 115
E D+ VE ++ +++E V + K S+ E+E++ N + K A A
Sbjct: 509 ENADDAARAVESLNGKKVDDKEWYVGKAQKKSEREQELK----NKFEQSMKEA---ADKY 667
Query: 116 HGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQA 175
G+ +Y+ + +++++E L+ S G ++ ++M+ +G +G FVAF T E AS+A
Sbjct: 668 QGANLYVKNLDDSIADEKLKELFSSYGTITSCKVMR-DPNGISRGSGFVAFSTPEEASRA 844
Query: 176 IEELNNSEFKGKKIKCSSSQAK 197
+ E+N K + + +Q K
Sbjct: 845 LLEMNGKMVVSKPLYVTLAQRK 910
>BP047377
Length = 564
Score = 52.8 bits (125), Expect = 7e-08
Identities = 25/71 (35%), Positives = 40/71 (56%)
Frame = +3
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
V++G I + +EE L CQ VG V R++ +E+G+ KGY F +K +E A A L
Sbjct: 135 VFVGNIPYDATEEQLIEICQEVGPVVSFRLVIDRETGKPKGYGFCEYKDEETALSARRNL 314
Query: 180 NNSEFKGKKIK 190
E G++++
Sbjct: 315 QGYEINGRQLR 347
Score = 46.2 bits (108), Expect = 6e-06
Identities = 26/71 (36%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Frame = +3
Query: 192 SSSQAKHR-LFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYN 250
+SSQ++HR +F+GNIP T E + ++ +VGP V+S L+ D + +G+ +G+ F EY +
Sbjct: 108 ASSQSQHRCVFVGNIPYDATEEQLIEICQEVGP-VVSFRLVID-RETGKPKGYGFCEYKD 281
Query: 251 HACAEYSRQKM 261
A +R+ +
Sbjct: 282 EETALSARRNL 314
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 51.6 bits (122), Expect = 2e-07
Identities = 23/66 (34%), Positives = 44/66 (65%)
Frame = +3
Query: 20 PEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEE 79
P+G P KP +D + + D D ++++E E ++ +++++ + EE E+ ++++EEEE EE
Sbjct: 216 PDG---PPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEE 386
Query: 80 VEEESK 85
VE E K
Sbjct: 387 VEVEGK 404
Score = 44.3 bits (103), Expect = 2e-05
Identities = 19/54 (35%), Positives = 39/54 (72%)
Frame = +3
Query: 43 DQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVD 96
D EEE E+ +EEE+ EDE+ ++ ++++++ EEEE+ +++ + +G EE+ V+
Sbjct: 246 DNEEE---EDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVE 398
Score = 42.7 bits (99), Expect = 7e-05
Identities = 20/67 (29%), Positives = 37/67 (54%)
Frame = +3
Query: 11 DVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEE 70
++ P P E+ D +E+ D D D+D +++ + + EEE++ +D+E EE EE
Sbjct: 207 EIYPDGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEE 386
Query: 71 EEEEEEE 77
E E ++
Sbjct: 387 VEVEGKD 407
Score = 41.6 bits (96), Expect = 2e-04
Identities = 18/50 (36%), Positives = 32/50 (64%)
Frame = +3
Query: 61 DEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAEL 110
D E EE ++EEE++E+E++ +++ DGEEE D ++E E + E+
Sbjct: 246 DNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEV 395
Score = 39.3 bits (90), Expect = 8e-04
Identities = 17/55 (30%), Positives = 35/55 (62%)
Frame = +3
Query: 55 EEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAE 109
EEEE +DEE ++ +E++++++++++ EE + D ++E EVE K A+
Sbjct: 252 EEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDGAK 416
Score = 38.9 bits (89), Expect = 0.001
Identities = 20/61 (32%), Positives = 32/61 (51%)
Frame = +3
Query: 5 PGTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEV 64
P + V N E + + E + + D+ D D D+ +EEE + ++ EEEE E+ EV
Sbjct: 216 PDGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEV 395
Query: 65 E 65
E
Sbjct: 396 E 398
>TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (46%)
Length = 1150
Score = 50.4 bits (119), Expect = 3e-07
Identities = 37/127 (29%), Positives = 61/127 (47%), Gaps = 4/127 (3%)
Frame = +3
Query: 70 EEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNV 129
EE+ + + V + SDG + D+ N+E + P + VY+G + ++V
Sbjct: 243 EEKNSDNQNAVVLTNGSSDGGQ----DNNNEEAPENN--------PSYTTVYVGNLPHDV 386
Query: 130 SEEDLRVFCQ----SVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFK 185
++ +L CQ G + EVRI + KG+ FV + T E A+ AI+ N +
Sbjct: 387 TQAELH--CQFHALGAGVIEEVRIQR------DKGFGFVRYNTHEEAALAIQVANGRIVR 542
Query: 186 GKKIKCS 192
GK +KCS
Sbjct: 543 GKTMKCS 563
Score = 40.4 bits (93), Expect = 4e-04
Identities = 32/149 (21%), Positives = 59/149 (39%), Gaps = 42/149 (28%)
Frame = +3
Query: 145 SEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKC---------SSSQ 195
S+ R+M ++G KGY FV+F+ + A +I ++ ++I+C SSS+
Sbjct: 66 SDARVMWDHKTGRSKGYGFVSFRDHQDAQSSINDMTGKWLGNRQIRCNWATKGAGGSSSE 245
Query: 196 AKH---------------------------------RLFIGNIPKKWTVEDMKKVVADVG 222
K+ +++GN+P T ++ +G
Sbjct: 246 EKNSDNQNAVVLTNGSSDGGQDNNNEEAPENNPSYTTVYVGNLPHDVTQAELHCQFHALG 425
Query: 223 PGVISIELLKDLQSSGRNRGFAFIEYYNH 251
GVI ++ R++GF F+ Y H
Sbjct: 426 AGVIEEVRIQ------RDKGFGFVRYNTH 494
>BP036655
Length = 556
Score = 49.7 bits (117), Expect = 6e-07
Identities = 25/73 (34%), Positives = 40/73 (54%)
Frame = -3
Query: 34 EQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEM 93
E+ D D +E ++ EE EE +D+++ + E ++E EEE E E K + EEE
Sbjct: 410 EEEGLSDDGDDDEVVDEEEEVVEEWSDDDDINDDESDDEVEEESETEIETEKEEEEEEEC 231
Query: 94 RVDHTNDEVEKKK 106
V++ DE+ KK
Sbjct: 230 GVENVEDEMGVKK 192
Score = 47.8 bits (112), Expect = 2e-06
Identities = 30/74 (40%), Positives = 41/74 (54%), Gaps = 7/74 (9%)
Frame = -3
Query: 33 DEQVDFDGDNDQEEEIEYEEVEE-------EEEVEDEEVEEVEEEEEEEEEEEEVEEESK 85
+E + DGD+D+ + E E VEE ++ D+EVEE E E E E+EEE EEE
Sbjct: 407 EEGLSDDGDDDEVVDEEEEVVEEWSDDDDINDDESDDEVEEESETEIETEKEEEEEEECG 228
Query: 86 PSDGEEEMRVDHTN 99
+ E+EM V N
Sbjct: 227 VENVEDEMGVKKFN 186
Score = 41.2 bits (95), Expect = 2e-04
Identities = 26/68 (38%), Positives = 39/68 (57%), Gaps = 2/68 (2%)
Frame = -3
Query: 18 ESPEGNSEPEKPTDSDEQV--DFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEE 75
E + + ++ D +E+V ++ D+D ++ +EVEEE E E E E EEEEEEE
Sbjct: 404 EGLSDDGDDDEVVDEEEEVVEEWSDDDDINDDESDDEVEEESETEIET--EKEEEEEEEC 231
Query: 76 EEEEVEEE 83
E VE+E
Sbjct: 230 GVENVEDE 207
Score = 33.9 bits (76), Expect = 0.033
Identities = 33/111 (29%), Positives = 51/111 (45%), Gaps = 4/111 (3%)
Frame = -3
Query: 55 EEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALP 114
EEE + D+ +++E +EEEE VEE S ++++ D ++DEVE++ E+
Sbjct: 410 EEEGLSDDG----DDDEVVDEEEEVVEEWS----DDDDINDDESDDEVEEESETEIETEK 255
Query: 115 PHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGE----GKGY 161
E G NV +E +G R + KESG G GY
Sbjct: 254 EEEEEEECG--VENVEDE--------MGVKKFNRPSRCKESGNRDIFGDGY 132
>BI419822
Length = 380
Score = 49.7 bits (117), Expect = 6e-07
Identities = 21/77 (27%), Positives = 41/77 (52%)
Frame = +2
Query: 121 YIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELN 180
++GG+A E L GE+ E +++ +E+G +G+ FV F +++ AIE +N
Sbjct: 86 FVGGLAWATDNEALEKAFSPYGEIVESKVINDRETGRSRGFGFVTFASEQAMKDAIEAMN 265
Query: 181 NSEFKGKKIKCSSSQAK 197
G+ I + +Q++
Sbjct: 266 GQNLDGRNITVNEAQSR 316
>TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG
polyadenylate-binding protein {Saccharomyces
cerevisiae;} , partial (18%)
Length = 518
Score = 48.1 bits (113), Expect = 2e-06
Identities = 23/60 (38%), Positives = 38/60 (63%)
Frame = +3
Query: 117 GSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAI 176
GS +Y+ I +NVS+E+LR + G ++ ++MK E G +G+ FV F T E A++A+
Sbjct: 150 GSNIYVKNIDDNVSDEELRGHFSACGSITYAKVMK-DEKGISQGFGFVCFSTPEEANKAV 326
>TC10057 similar to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (49%)
Length = 598
Score = 48.1 bits (113), Expect = 2e-06
Identities = 31/136 (22%), Positives = 59/136 (42%), Gaps = 11/136 (8%)
Frame = +2
Query: 77 EEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRV 136
+EEV ++ DG E +V+ E E +++Y G + +V L
Sbjct: 215 QEEVALAAEADDGLVEEKVEEVVAEEEANSSVN--------TKLYFGNLPYSVDSAQLAG 370
Query: 137 FCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSS-- 194
+ G + ++ ++SG+ +G+AFV E + IE L+ EF G+ ++ + S
Sbjct: 371 LIEEYGSAELIEVLYDRDSGKSRGFAFVTMSCVEDCNTVIENLDGKEFLGRTLRVNLSDK 550
Query: 195 ---------QAKHRLF 201
+ +H+LF
Sbjct: 551 PKPKEPLYPETEHKLF 598
Score = 33.1 bits (74), Expect = 0.057
Identities = 38/150 (25%), Positives = 67/150 (44%)
Frame = +2
Query: 97 HTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESG 156
+TN + + K+ +LA V I +S L V S V++ + A E+
Sbjct: 71 NTNVKCLRSKNCSVLASVSQSYLVEPLSIGGTISHNKLWVPRISAA-VAQEEVALAAEAD 247
Query: 157 EGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKK 216
+G V K +E+ ++ EE N+S +L+ GN+P +
Sbjct: 248 DG----LVEEKVEEVVAE--EEANSS-------------VNTKLYFGNLPYSVDSAQLAG 370
Query: 217 VVADVGPGVISIELLKDLQSSGRNRGFAFI 246
++ + G + IE+L D + SG++RGFAF+
Sbjct: 371 LIEEYGSAEL-IEVLYD-RDSGKSRGFAFV 454
>BP085186
Length = 388
Score = 48.1 bits (113), Expect = 2e-06
Identities = 24/48 (50%), Positives = 30/48 (62%)
Frame = -1
Query: 39 DGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKP 86
DG+ ++EEE E EE EEEEE E+EE E EE+ EEE + E P
Sbjct: 274 DGEGNEEEEEEEEEEEEEEEEEEEEAEPEEEKNEEENVNTQEGEGENP 131
Score = 47.4 bits (111), Expect = 3e-06
Identities = 25/44 (56%), Positives = 28/44 (62%)
Frame = -1
Query: 49 EYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEE 92
E E EEEEE E+EE EE EEEE E EEE+ EE +GE E
Sbjct: 268 EGNEEEEEEEEEEEEEEEEEEEEAEPEEEKNEEENVNTQEGEGE 137
Score = 45.1 bits (105), Expect = 1e-05
Identities = 25/53 (47%), Positives = 34/53 (63%)
Frame = -1
Query: 32 SDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEES 84
S + D +G+ ++EEE E EE EEEEE E+ E EE E+ EEE +E E E+
Sbjct: 289 SPDNADGEGNEEEEEEEEEEEEEEEEEEEEAEPEE-EKNEEENVNTQEGEGEN 134
Score = 44.7 bits (104), Expect = 2e-05
Identities = 22/60 (36%), Positives = 34/60 (56%)
Frame = -1
Query: 33 DEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEE 92
D ++ +G N + + + + + EE EEEEEEEEEEEE EEE++P + + E
Sbjct: 352 DLAINLEGSNPYARIVNGKLISPDNADGEGNEEEEEEEEEEEEEEEEEEEEAEPEEEKNE 173
Score = 39.3 bits (90), Expect = 8e-04
Identities = 24/69 (34%), Positives = 38/69 (54%)
Frame = -1
Query: 11 DVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEE 70
++ NP + N + P ++D + + + + ++EEE E EE EEEEE E EE + EE
Sbjct: 340 NLEGSNPYARIVNGKLISPDNADGEGNEEEEEEEEEEEEEEE-EEEEEAEPEEEKNEEEN 164
Query: 71 EEEEEEEEE 79
+E E E
Sbjct: 163 VNTQEGEGE 137
Score = 27.7 bits (60), Expect = 2.4
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = -1
Query: 72 EEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKK 105
E EEEEE EEE + + EEE + ++ E++
Sbjct: 268 EGNEEEEEEEEEEEEEEEEEEEEAEPEEEKNEEE 167
Score = 25.8 bits (55), Expect = 9.0
Identities = 12/40 (30%), Positives = 19/40 (47%)
Frame = -1
Query: 67 VEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKK 106
+ + + E EE EEE + + EEE + E EK +
Sbjct: 292 ISPDNADGEGNEEEEEEEEEEEEEEEEEEEEAEPEEEKNE 173
>TC16073 similar to UP|Q9FJN9 (Q9FJN9) Poly(A)-binding protein II-like,
partial (59%)
Length = 529
Score = 46.6 bits (109), Expect = 5e-06
Identities = 44/162 (27%), Positives = 72/162 (44%), Gaps = 18/162 (11%)
Frame = +1
Query: 43 DQEEEIEYEEVEEEEEVE-----DEEVEEVEEE----------EEEEEEEEEVEEES--- 84
DQEE Y +EEVE D + E+ + + E+ ++ +E+EEE+
Sbjct: 55 DQEEHEVYGADIPDEEVEMDADADADAEQQDHDFASNHPNKDLEDMKKRLKEIEEEASAL 234
Query: 85 KPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEV 144
+ + E + D AE + G +Y+G + + E+++ QS G V
Sbjct: 235 REMQAKVEKDMGAVQDAGSSATQAEKEEV--DGRSIYVGNVDYACTPEEVQQHFQSCGTV 408
Query: 145 SEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKG 186
+ V I+ K G+ KG+A+V F E A+ LN SE G
Sbjct: 409 NRVTILTDK-FGQPKGFAYVEFVEAEAVQNAV-MLNESELHG 528
>BP031757
Length = 507
Score = 46.2 bits (108), Expect = 6e-06
Identities = 28/88 (31%), Positives = 42/88 (46%)
Frame = +2
Query: 3 KMPGTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDE 62
KM R V + E E + KP + DN+ ++ +YEE EEE + +
Sbjct: 56 KMVNRRFTQVATSDDEEEETPPQQLKPRKRKRMKLLEEDNEDKDGEDYEE---EEEKKKK 226
Query: 63 EVEEVEEEEEEEEEEEEVEEESKPSDGE 90
+ ++EEEEEEE+EE E P D +
Sbjct: 227 KKSRKKKEEEEEEEQEEPESPQPPEDAK 310
Score = 31.6 bits (70), Expect = 0.16
Identities = 18/69 (26%), Positives = 39/69 (56%), Gaps = 1/69 (1%)
Frame = +2
Query: 42 NDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEE-EEEEVEEESKPSDGEEEMRVDHTND 100
+D+EEE ++++ + + +EE E+++ E+ EEEE +++ K S ++E + +
Sbjct: 95 DDEEEETPPQQLKPRKRKRMKLLEEDNEDKDGEDYEEEEEKKKKKKSRKKKEEEEEEEQE 274
Query: 101 EVEKKKHAE 109
E E + E
Sbjct: 275 EPESPQPPE 301
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 46.2 bits (108), Expect = 6e-06
Identities = 32/82 (39%), Positives = 44/82 (53%), Gaps = 6/82 (7%)
Frame = +3
Query: 15 QNPESPEGNSEPEK-----PTDSDEQVDFDGDNDQEEEIEYEEVEEEEEV-EDEEVEEVE 68
Q E E + E E+ PT +V+ + D+EEE EE EEEE V EDE+ E E
Sbjct: 330 QGVEEEEEDEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGEEEEEEEEVVVEDEQGNEDE 509
Query: 69 EEEEEEEEEEEVEEESKPSDGE 90
+E+EEEE VE + + D +
Sbjct: 510 DEDEEEEGVVVVEVKGRKVDSK 575
Score = 43.1 bits (100), Expect = 5e-05
Identities = 26/64 (40%), Positives = 35/64 (54%), Gaps = 10/64 (15%)
Frame = +3
Query: 51 EEVEEEEEVEDEE----------VEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTND 100
EE EE+EE E+E+ VE +EE+EEEE EE EEE + +E+ D D
Sbjct: 339 EEEEEDEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGEEEEEEEEVVVEDEQGNEDEDED 518
Query: 101 EVEK 104
E E+
Sbjct: 519 EEEE 530
Score = 41.2 bits (95), Expect = 2e-04
Identities = 27/59 (45%), Positives = 33/59 (55%), Gaps = 11/59 (18%)
Frame = +3
Query: 45 EEEIEYEEVEEEE-----------EVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEE 92
EEE E EE EEE+ E EE EE E EEEEEEEE V E+ + ++ E+E
Sbjct: 339 EEEEEDEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGEEEEEEEEVVVEDEQGNEDEDE 515
Score = 40.8 bits (94), Expect = 3e-04
Identities = 19/46 (41%), Positives = 29/46 (62%)
Frame = +3
Query: 64 VEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAE 109
VE++ + EEEEE+EE EEE P RV+H +E E++++ E
Sbjct: 315 VEKLNQGVEEEEEDEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGE 452
Score = 33.5 bits (75), Expect = 0.043
Identities = 26/84 (30%), Positives = 42/84 (49%), Gaps = 11/84 (13%)
Frame = +3
Query: 53 VEEEEEVEDEEVEEVEE--------EEEEEEEEEEVEEESKPSDGEEEMRVDH---TNDE 101
VEEEEE E++E E+ E +EE+EE EE + + EEE+ V+ DE
Sbjct: 336 VEEEEEDEEDEEEQTPHPTRGRSRVEHAQEEDEE--EENGEEEEEEEEVVVEDEQGNEDE 509
Query: 102 VEKKKHAELLALPPHGSEVYIGGI 125
E ++ ++ + G +V G+
Sbjct: 510 DEDEEEEGVVVVEVKGRKVDSKGL 581
Score = 30.0 bits (66), Expect = 0.48
Identities = 15/42 (35%), Positives = 26/42 (61%)
Frame = +2
Query: 48 IEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDG 89
I+Y + E+E+E EDE+ ++E E+EE + E++P G
Sbjct: 227 IDYLDEEDEDEDEDED----DDERGREKEEAQAG*EAEPGGG 340
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.303 0.125 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,010,302
Number of Sequences: 28460
Number of extensions: 52007
Number of successful extensions: 3460
Number of sequences better than 10.0: 359
Number of HSP's better than 10.0 without gapping: 810
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2006
length of query: 289
length of database: 4,897,600
effective HSP length: 89
effective length of query: 200
effective length of database: 2,364,660
effective search space: 472932000
effective search space used: 472932000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0010.10


