

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
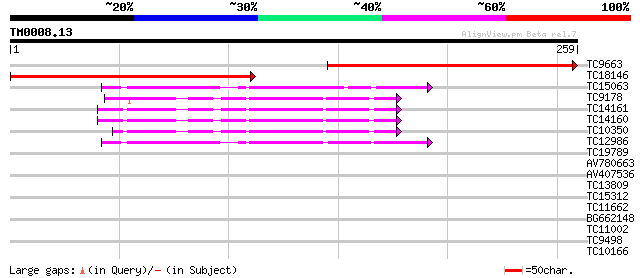
Query= TM0008.13
(259 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9663 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle... 228 8e-61
TC18146 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particl... 224 1e-59
TC15063 homologue to UP|SARA_ARATH (O04834) GTP-binding protein ... 45 1e-05
TC9178 homologue to GB|AAB17725.1|1654142|BCU38470 small GTP-bin... 44 2e-05
TC14161 UP|AAR29293 (AAR29293) ADP-ribosylation factor, complete 43 5e-05
TC14160 homologue to GB|BAB90396.1|20161472|AP003570 ADP-ribosyl... 43 5e-05
TC10350 homologue to UP|AAR29293 (AAR29293) ADP-ribosylation fac... 43 6e-05
TC12986 homologue to UP|SARB_ARATH (Q01474) GTP-binding protein ... 42 8e-05
TC19789 38 0.002
AV780663 35 0.017
AV407536 29 0.92
TC13809 28 1.6
TC15312 UP|RPOD_LOTJA (Q9BBS7) DNA-directed RNA polymerase beta'... 28 2.1
TC11662 similar to UP|O65748 (O65748) Acyl-CoA synthetase (Frag... 26 7.8
BG662148 26 7.8
TC11002 similar to GB|AAD39677.1|5103847|F9L1 Contains PF|00561 ... 26 7.8
TC9498 26 7.8
TC10166 weakly similar to UP|Q8W0T8 (Q8W0T8) Gb protein, partial... 26 7.8
>TC9663 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (44%)
Length = 612
Score = 228 bits (581), Expect = 8e-61
Identities = 114/114 (100%), Positives = 114/114 (100%)
Frame = +1
Query: 146 PNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRS 205
PNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRS
Sbjct: 1 PNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLEKEIDKLRASRS 180
Query: 206 AVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFIREYVKP 259
AVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFIREYVKP
Sbjct: 181 AVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEEFIREYVKP 342
>TC18146 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (28%)
Length = 463
Score = 224 bits (571), Expect = 1e-59
Identities = 112/112 (100%), Positives = 112/112 (100%)
Frame = +2
Query: 1 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 60
MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL
Sbjct: 128 MQELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVL 307
Query: 61 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHI 112
TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHI
Sbjct: 308 TGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHI 463
>TC15063 homologue to UP|SARA_ARATH (O04834) GTP-binding protein SAR1A,
complete
Length = 933
Score = 45.4 bits (106), Expect = 1e-05
Identities = 46/151 (30%), Positives = 69/151 (45%)
Frame = +2
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 194 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQYPTSEELSI------ 349
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y + VV++VDA D + E L +L+
Sbjct: 350 --GKIK-FKAFDLGGHQIARRVWKDYYAKVDAVVYLVDAYDKERFAESKKE-LDALLSDE 517
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P LIL NK D A ++E +R L
Sbjct: 518 SLA--NVPFLILGNKIDIPYAASEEELRYSL 604
>TC9178 homologue to GB|AAB17725.1|1654142|BCU38470 small GTP-binding
protein ARF {Brassica rapa subsp. pekinensis;} ,
complete
Length = 1033
Score = 44.3 bits (103), Expect = 2e-05
Identities = 40/139 (28%), Positives = 63/139 (44%), Gaps = 3/139 (2%)
Frame = +3
Query: 44 LLSLRLFKRA---KTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
L+ RLF K +++ GL +GKT + Y+L+ G V S P G + E
Sbjct: 267 LVFTRLFSSVFGNKEARILVLGLDNAGKTTILYRLQ-----MGEVVSTIPTIG---FNVE 422
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + +K + D+ G + +RP Y P +++VVD+ D A E + IL
Sbjct: 423 TVQYNNIK-FQVWDLGGQTSIRPYWRCYFPNTQAIIYVVDSSD-TDRLVIAKEEFHAILE 596
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + K +LI NK D
Sbjct: 597 EEEL--KGAVVLIFANKQD 647
>TC14161 UP|AAR29293 (AAR29293) ADP-ribosylation factor, complete
Length = 912
Score = 43.1 bits (100), Expect = 5e-05
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +2
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 98 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 250
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 251 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 424
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 425 EDEL--RDAVLLVFANKQD 475
>TC14160 homologue to GB|BAB90396.1|20161472|AP003570 ADP-ribosylation
factor {Oryza sativa (japonica cultivar-group);},
partial (48%)
Length = 1105
Score = 43.1 bits (100), Expect = 5e-05
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +3
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 216 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 368
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 369 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 542
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 543 EDEL--RDAVLLVFANKQD 593
>TC10350 homologue to UP|AAR29293 (AAR29293) ADP-ribosylation factor,
partial (98%)
Length = 603
Score = 42.7 bits (99), Expect = 6e-05
Identities = 36/132 (27%), Positives = 62/132 (46%)
Frame = +3
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 102 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 254
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 255 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRAVEARDELHRMLNEDEL--R 422
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 423 DAVLLVFANKQD 458
>TC12986 homologue to UP|SARB_ARATH (Q01474) GTP-binding protein SAR1B,
partial (80%)
Length = 515
Score = 42.4 bits (98), Expect = 8e-05
Identities = 43/151 (28%), Positives = 66/151 (43%)
Frame = +1
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 79 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI------ 234
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y + V ++VDA D A S+ D L
Sbjct: 235 --GKIK-FKAFDLGGHQVARRVWKDYYAKVDAVAYLVDAYD--KERFAESKKELDALLSD 399
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
+ +P L+L NK D A +++ +R L
Sbjct: 400 EAL-ASVPFLVLGNKIDIPYASSEDELRYHL 489
>TC19789
Length = 525
Score = 38.1 bits (87), Expect = 0.002
Identities = 38/146 (26%), Positives = 68/146 (46%)
Frame = +3
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
R+FK+ +T G+ GKT +L +G+ ++++ +P E TF+ ETT K
Sbjct: 102 RMFKKTQTVQACFLGIASPGKTSFAQRLSEGNK---SMSACDPTE-TFL---ETTATHKK 260
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ + ++ LR + Q V+ VDA + P+ +AS K + ++
Sbjct: 261 HLIVMREIGSKETLRNLWKHEIQQTNVFVWFVDASN--PDSWSASSEALLTFIKHNALEH 434
Query: 168 KIPLLILCNKTDKVTAHTKEFIRRQL 193
+ P LIL K DK A + + + + L
Sbjct: 435 Q-PWLILVTKVDKAEAKSVDEVVKAL 509
>AV780663
Length = 568
Score = 34.7 bits (78), Expect = 0.017
Identities = 19/55 (34%), Positives = 28/55 (50%)
Frame = +1
Query: 87 SMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDA 141
S+E G I TT K +++VD PGH+ L +++ + G V VVDA
Sbjct: 280 SLERERGITIASKVTTVSWKENELNMVDTPGHADLGGEVERVVGMVEGAV*VVDA 444
>AV407536
Length = 382
Score = 28.9 bits (63), Expect = 0.92
Identities = 31/130 (23%), Positives = 48/130 (36%), Gaps = 15/130 (11%)
Frame = +2
Query: 66 SGKT-----VLFY----------QLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPV 110
SGKT VLFY + RDG + +E +G I + T K +
Sbjct: 17 SGKTTLTERVLFYAGRIHEIHEVRGRDGVGAKMDSMDLEREKGITIQSAATYCSWKDCKI 196
Query: 111 HIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIP 170
+I+D PGH +++ L G + V+ C I + + +P
Sbjct: 197 NIIDTPGHVDFTIEVERALRVLDGAILVL--------CSVGGVQSQSITVDRQMRRYDVP 352
Query: 171 LLILCNKTDK 180
L NK D+
Sbjct: 353 RLAFINKLDR 382
>TC13809
Length = 528
Score = 28.1 bits (61), Expect = 1.6
Identities = 13/39 (33%), Positives = 23/39 (58%)
Frame = +1
Query: 74 QLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHI 112
+++D + T+T + PNEGT + + TK G +K H+
Sbjct: 253 EVKDKTRFVSTITMIMPNEGT--IDMQRTKLGVIKRAHM 363
>TC15312 UP|RPOD_LOTJA (Q9BBS7) DNA-directed RNA polymerase beta'' chain
(PEP) (Plastid-encoded RNA polymerase beta'' subunit)
(RNA polymerase beta'' subunit) , partial (84%)
Length = 3522
Score = 27.7 bits (60), Expect = 2.1
Identities = 13/51 (25%), Positives = 25/51 (48%)
Frame = -3
Query: 23 RQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFY 73
R PP +L++ I VFT ++LL + + G+ + T+++Y
Sbjct: 1423 RTNPPTRLFLFKVIGVFTPMILLFRTIMEEDSDKICTSWGMKKNRSTLIWY 1271
>TC11662 similar to UP|O65748 (O65748) Acyl-CoA synthetase (Fragment) ,
partial (81%)
Length = 640
Score = 25.8 bits (55), Expect = 7.8
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = -2
Query: 164 VVKKKIPLLILCNKTDKVTAHTK 186
V KK PLL++C +DKVT H K
Sbjct: 342 VFKKLRPLLLVCG-SDKVTFHVK 277
>BG662148
Length = 415
Score = 25.8 bits (55), Expect = 7.8
Identities = 13/39 (33%), Positives = 20/39 (50%)
Frame = +3
Query: 75 LRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIV 113
L+D + H T+ +P EGT + S KV +HI+
Sbjct: 171 LKDQNHHMETLIFTDPLEGTLMAESSLIS*HKVLDLHIL 287
>TC11002 similar to GB|AAD39677.1|5103847|F9L1 Contains PF|00561
alpha/beta hydrolase fold. {Arabidopsis thaliana;} ,
partial (34%)
Length = 1021
Score = 25.8 bits (55), Expect = 7.8
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = +1
Query: 26 PPEQLYIAAAIAVFTTLLLLSLRLFKR 52
PPE IA A+AV +LLL + F+R
Sbjct: 49 PPEPSIIAVAVAVVHGVLLLPKQSFRR 129
>TC9498
Length = 466
Score = 25.8 bits (55), Expect = 7.8
Identities = 16/35 (45%), Positives = 19/35 (53%)
Frame = +2
Query: 217 TLGVPGEAFSFTQCCNKVTTADASGLTGEISQLEE 251
TL + G AF F CC+ + T S G ISQ EE
Sbjct: 281 TLALLGAAF-FVVCCSAIYTRLCSRNRGIISQREE 382
>TC10166 weakly similar to UP|Q8W0T8 (Q8W0T8) Gb protein, partial (5%)
Length = 980
Score = 25.8 bits (55), Expect = 7.8
Identities = 17/46 (36%), Positives = 23/46 (49%)
Frame = -1
Query: 26 PPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVLTGLTGSGKTVL 71
PP LY +A+ V + +LLS L K +L GL G +T L
Sbjct: 515 PPTWLYRFSAVKVLLSFVLLSFDLVK-CVLKEELLRGLVGIHRTFL 381
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,162,457
Number of Sequences: 28460
Number of extensions: 50472
Number of successful extensions: 254
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 252
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 254
length of query: 259
length of database: 4,897,600
effective HSP length: 88
effective length of query: 171
effective length of database: 2,393,120
effective search space: 409223520
effective search space used: 409223520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0008.13


