

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
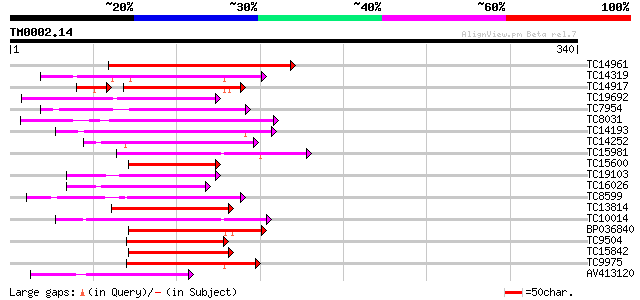
Query= TM0002.14
(340 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14961 similar to UP|Q9LEM7 (Q9LEM7) AP2-domain DNA-binding pro... 105 8e-24
TC14319 similar to UP|AP23_ARATH (P42736) AP2 domain transcripti... 99 1e-21
TC14917 similar to GB|AAP37839.1|30725634|BT008480 At5g64750 {Ar... 89 2e-20
TC19692 similar to UP|Q9LYD3 (Q9LYD3) Transcription factor like ... 94 3e-20
TC7954 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protein... 90 6e-19
TC8031 similar to UP|Q9FR33 (Q9FR33) Ripening regulated protein ... 87 5e-18
TC14193 similar to UP|Q8S2S7 (Q8S2S7) Ethylene responsive elemen... 86 9e-18
TC14252 homologue to UP|Q9ZR85 (Q9ZR85) Ethylene-responsive elem... 86 1e-17
TC15981 similar to UP|ERF3_ARATH (O80339) Ethylene responsive el... 84 3e-17
TC15600 homologue to UP|BAD01556 (BAD01556) ERF-like protein, pa... 84 3e-17
TC19103 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive e... 84 3e-17
TC16026 similar to UP|Q9SGZ3 (Q9SGZ3) F28K19.29, partial (32%) 82 1e-16
TC8599 homologue to UP|Q40478 (Q40478) EREBP-4, partial (26%) 82 1e-16
TC13814 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protei... 82 1e-16
TC10014 similar to UP|Q8LLR3 (Q8LLR3) Ethylene-responsive elemen... 82 2e-16
BP036840 82 2e-16
TC9504 similar to UP|Q9SXS8 (Q9SXS8) Ethylene responsive element... 80 5e-16
TC15842 homologue to GB|BAB62912.1|15207792|AB055883 ERF domain ... 80 6e-16
TC9975 similar to UP|AAR15448 (AAR15448) AP2 transcription facto... 78 2e-15
AV413120 75 1e-14
>TC14961 similar to UP|Q9LEM7 (Q9LEM7) AP2-domain DNA-binding protein,
partial (27%)
Length = 1135
Score = 105 bits (263), Expect = 8e-24
Identities = 55/112 (49%), Positives = 68/112 (60%)
Frame = +1
Query: 60 KGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILY 119
KGKGGP+N++ YRGVRQR+WGKWVAEIREP + R WLGTF+TA AA AYD AA +Y
Sbjct: 352 KGKGGPENSRCNYRGVRQRTWGKWVAEIREPNRGKRLWLGTFATAIGAALAYDEAARAMY 531
Query: 120 GSRAQLNLQPSPTSSSSQSSSSSSRASSSSSSTQTLRPLLPRPSGYGFSFSG 171
GS A+LN S S+ S S ++ S + L S + SG
Sbjct: 532 GSSARLNFPNVAVSHFSEESLKDSSVANDDSGSSMAAVLANTESKVSSNNSG 687
>TC14319 similar to UP|AP23_ARATH (P42736) AP2 domain transcription factor
RAP2.3 (Related to AP2 protein 3) (Cadmium-induced
protein AS30), partial (37%)
Length = 1130
Score = 99.0 bits (245), Expect = 1e-21
Identities = 63/150 (42%), Positives = 81/150 (54%), Gaps = 14/150 (9%)
Frame = +3
Query: 19 TATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCK------GKGGPDNNKFR- 71
+A AA++TTP+ S+S N ++ I P+ ++K + G GG R
Sbjct: 222 SAGAAVSTTPTSKSSSPPLN--FDDVIQVPDQKVAGCDKKGEKKKSVVGSGGAGKKNARK 395
Query: 72 --YRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNL-- 127
YRG+RQR WGKW AEIR+P+K R WLGTF TAE+AARAYD AA + G +A+LN
Sbjct: 396 NVYRGIRQRPWGKWAAEIRDPQKGVRVWLGTFPTAEEAARAYDEAAKRIRGDKAKLNFPD 575
Query: 128 ---QPSPTSSSSQSSSSSSRASSSSSSTQT 154
P PT S SSS SS T
Sbjct: 576 TVAAPPPTKKQRCLSPEQPEYSSSGSSGPT 665
>TC14917 similar to GB|AAP37839.1|30725634|BT008480 At5g64750 {Arabidopsis
thaliana;}, partial (17%)
Length = 1051
Score = 89.4 bits (220), Expect(2) = 2e-20
Identities = 47/84 (55%), Positives = 59/84 (69%), Gaps = 11/84 (13%)
Frame = +3
Query: 69 KFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNL- 127
K +YRGVRQR WGKW AEIR+P K R WLGTF+TAE+AARAYD+A++ G++A+LN
Sbjct: 201 KRKYRGVRQRPWGKWAAEIRDPFKAARVWLGTFNTAEEAARAYDQASLRFRGNKAKLNFP 380
Query: 128 -------QPS---PTSSSSQSSSS 141
QPS PT+ S S+SS
Sbjct: 381 ENVRLLQQPSINQPTTHFSNSNSS 452
Score = 30.8 bits (68), Expect = 0.34
Identities = 29/88 (32%), Positives = 41/88 (45%), Gaps = 2/88 (2%)
Frame = +3
Query: 102 STAEDAARAYDRAAIILYGSRAQLNLQPSP--TSSSSQSSSSSSRASSSSSSTQTLRPLL 159
S+ +++ YD + + LY + S SSSS SSS SS +SS SS Q +
Sbjct: 498 SSGSNSSNFYDYSPMSLYDQANVSSTMASHHLQSSSSSSSSYSSMFASSVSSLQEVSV-- 671
Query: 160 PRPSGYGFSFSGSHLPVVFSAAASGPFG 187
PS Y + LP +S +S P G
Sbjct: 672 --PSVY-----STQLPAQYSVHSSSPSG 734
Score = 26.2 bits (56), Expect(2) = 2e-20
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 2/23 (8%)
Frame = +2
Query: 41 HNNNITTPN--TSTTINNRKCKG 61
H+NN PN TS I NR+C+G
Sbjct: 113 HSNNKNKPNGETSIRIQNRECEG 181
>TC19692 similar to UP|Q9LYD3 (Q9LYD3) Transcription factor like protein,
partial (29%)
Length = 454
Score = 94.0 bits (232), Expect = 3e-20
Identities = 51/120 (42%), Positives = 70/120 (57%), Gaps = 1/120 (0%)
Frame = +3
Query: 8 PHHPPQEITK-PTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPD 66
PH P K P ++ P + NSNS+ ++++ + + NN + +
Sbjct: 93 PHKPSYSNFKYPILFITMSVEPHNSETESNSNSNSYTSLSSSSPPQSNNNNTKRNR--ES 266
Query: 67 NNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLN 126
N YRGVR R+WGKWV+EIREPRK++R WLGTF T E AARA+D AA+ + GS A LN
Sbjct: 267 NTHAVYRGVRMRTWGKWVSEIREPRKKSRIWLGTFCTPEMAARAHDVAALSIKGSAAILN 446
>TC7954 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protein, partial
(75%)
Length = 1708
Score = 89.7 bits (221), Expect = 6e-19
Identities = 50/126 (39%), Positives = 74/126 (58%)
Frame = +2
Query: 19 TATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQR 78
+ATAA T + SNS + S+ ++ + + ++K K + YRG+RQR
Sbjct: 401 SATAAAKT---KRSNSFSKGSTAAKSVESNEQAEKYASKKRKNQ---------YRGIRQR 544
Query: 79 SWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQS 138
WGKW AEIR+PRK R WLGTF+TAE+AARAYD A + G++A++N P SS +
Sbjct: 545 PWGKWAAEIRDPRKGVRVWLGTFNTAEEAARAYDAEARRIRGNKAKVNFPVEPQLSSKRL 724
Query: 139 SSSSSR 144
++ +
Sbjct: 725 KANPEK 742
>TC8031 similar to UP|Q9FR33 (Q9FR33) Ripening regulated protein DDTFR10/A
(Fragment), partial (44%)
Length = 1109
Score = 86.7 bits (213), Expect = 5e-18
Identities = 56/157 (35%), Positives = 78/157 (49%), Gaps = 2/157 (1%)
Frame = +2
Query: 7 TPHHPPQEIT-KPTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGP 65
TP PPQ + KP+ A+ +E N + PN + + + P
Sbjct: 395 TPVAPPQRLARKPSLQIALPKKTTEWIQFGNPD---------PNPDPVV-----QAQPEP 532
Query: 66 DNNKFRYRGVRQRSWGKWVAEIREPRKR-TRKWLGTFSTAEDAARAYDRAAIILYGSRAQ 124
K YRGVRQR WGK+ AEIR+P KR +R WLGTF TA +AA+AYDRAA L GS+A
Sbjct: 533 AEKKQHYRGVRQRPWGKFAAEIRDPNKRGSRVWLGTFDTAIEAAKAYDRAAFKLRGSKAI 712
Query: 125 LNLQPSPTSSSSQSSSSSSRASSSSSSTQTLRPLLPR 161
LN ++++ S R ++ ++ R
Sbjct: 713 LNFPLDVAAAAASGDGESDRKRRREEEVVEVKAVVKR 823
>TC14193 similar to UP|Q8S2S7 (Q8S2S7) Ethylene responsive element binding
factor 4-like protein, partial (50%)
Length = 1128
Score = 85.9 bits (211), Expect = 9e-18
Identities = 48/137 (35%), Positives = 79/137 (57%), Gaps = 4/137 (2%)
Frame = +3
Query: 28 PSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEI 87
PS T+ ++ N++ TT +T ++ + K + + +RGVR+R WG++ AEI
Sbjct: 54 PSSTTVTKARNTAA---ATTAAATTAVSAKSKIHKPSNNATEVHFRGVRKRPWGRYAAEI 224
Query: 88 REPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSS----SSS 143
R+P K++R WLGTF TAE+AARAYD AA G +A+ N SP+ ++ + ++
Sbjct: 225 RDPGKKSRVWLGTFDTAEEAARAYDAAARQFRGPKAKTNFPQSPSPDNNTGPNDVVVANV 404
Query: 144 RASSSSSSTQTLRPLLP 160
+A+ S S + T+ P
Sbjct: 405 KANQSPSQSSTVESSTP 455
Score = 26.6 bits (57), Expect = 6.4
Identities = 10/25 (40%), Positives = 19/25 (76%), Gaps = 1/25 (4%)
Frame = +1
Query: 6 TTPHHPPQEIT-KPTATAAITTTPS 29
T+P+HPP+ T PT ++++T+ P+
Sbjct: 340 TSPNHPPRTTTPDPTTSSSLTSRPT 414
>TC14252 homologue to UP|Q9ZR85 (Q9ZR85) Ethylene-responsive element binding
protein homolog, partial (47%)
Length = 1174
Score = 85.5 bits (210), Expect = 1e-17
Identities = 49/109 (44%), Positives = 63/109 (56%), Gaps = 4/109 (3%)
Frame = +1
Query: 45 ITTPNTSTTINNRKCKGKGGPDNN----KFRYRGVRQRSWGKWVAEIREPRKRTRKWLGT 100
I P T N K G G + N + +RGVR+R WG++ AEIR+P K++R WLGT
Sbjct: 112 IMAPRDKT--NAVKVSGNGNGNGNGGVKEVHFRGVRKRPWGRYAAEIRDPGKKSRVWLGT 285
Query: 101 FSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRASSSS 149
F TAE+AARAYD AA G +A+ N + +QS S SS SSS
Sbjct: 286 FDTAEEAARAYDTAAREFRGPKAKTNFPFPSDNVKNQSPSQSSTVESSS 432
>TC15981 similar to UP|ERF3_ARATH (O80339) Ethylene responsive element
binding factor 3 (AtERF3), partial (45%)
Length = 671
Score = 84.3 bits (207), Expect = 3e-17
Identities = 50/121 (41%), Positives = 67/121 (55%), Gaps = 4/121 (3%)
Frame = +1
Query: 65 PDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQ 124
P+ + R+RGVR+R WG++ AEIR+P K+ R WLGTF +AEDAARAYD AA G +A+
Sbjct: 220 PETKETRFRGVRKRPWGRFAAEIRDPWKKARVWLGTFDSAEDAARAYDAAARSFRGPKAK 399
Query: 125 LNLQPSPTSSSSQSSSSSSRASSSS----SSTQTLRPLLPRPSGYGFSFSGSHLPVVFSA 180
N P+P + S+ S S +RP S SFSG +P +A
Sbjct: 400 TNF-PTPDEIIAGGGDEISQIPISHHHGFSKFDPVRPTTSGMSSTVESFSGPRVPSSSAA 576
Query: 181 A 181
A
Sbjct: 577 A 579
>TC15600 homologue to UP|BAD01556 (BAD01556) ERF-like protein, partial (36%)
Length = 570
Score = 84.3 bits (207), Expect = 3e-17
Identities = 37/55 (67%), Positives = 45/55 (81%)
Frame = +3
Query: 72 YRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLN 126
YRG+RQR WGKW AEIR+PRK R WLGTF+TAE+AARAYDR A + G +A++N
Sbjct: 396 YRGIRQRPWGKWAAEIRDPRKGVRVWLGTFNTAEEAARAYDREARKIRGKKAKVN 560
>TC19103 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive element
binding protein, partial (37%)
Length = 382
Score = 84.0 bits (206), Expect = 3e-17
Identities = 44/92 (47%), Positives = 55/92 (58%)
Frame = +1
Query: 35 ENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRT 94
E S N+ ITT + S+ N RK YRG+RQR WGKWVAE+REP KR+
Sbjct: 61 ERDCCSTNSTITTTSNSSRENKRK-------HQQDKPYRGIRQRKWGKWVAEVREPNKRS 219
Query: 95 RKWLGTFSTAEDAARAYDRAAIILYGSRAQLN 126
R WLG++++ AARAYD A L G A+LN
Sbjct: 220 RIWLGSYASPVAAARAYDTAVFYLRGPSARLN 315
>TC16026 similar to UP|Q9SGZ3 (Q9SGZ3) F28K19.29, partial (32%)
Length = 1183
Score = 82.4 bits (202), Expect = 1e-16
Identities = 45/86 (52%), Positives = 51/86 (58%)
Frame = +2
Query: 35 ENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRT 94
+N N +H+ N T S K P YRGVRQR WGKWVAEIR P+ RT
Sbjct: 926 QNQNQNHHLNTTLSFLSP--KPIPMKHVANPPKPTKLYRGVRQRHWGKWVAEIRLPKNRT 1099
Query: 95 RKWLGTFSTAEDAARAYDRAAIILYG 120
R WLGTF TAE+AA AYD+AA L G
Sbjct: 1100 RLWLGTFDTAEEAALAYDKAAYKLRG 1177
Score = 38.1 bits (87), Expect = 0.002
Identities = 40/131 (30%), Positives = 55/131 (41%), Gaps = 8/131 (6%)
Frame = +2
Query: 129 PSPTSSSSQSSSSSSRASSSSS-STQTLRPLLPRPSGYGFSFSGSHLPVVFSAAASGPFG 187
P+PTS+S+ S SSSS +SSSSS S + LP S +S + FS S
Sbjct: 608 PTPTSTSTFSPSSSSSSSSSSSPSPSSSNSSLPCSSPPNSFYSSPSVTPFFSDGCSTSMT 787
Query: 188 FYNANNGYMQLQPQQ-------HHFHHQEVVQSQLQPQLQQCRQPEPDVKGGGVVDHVRS 240
A NG +QP +H ++ Q Q Q Q+QQ
Sbjct: 788 HVFA-NGLSFVQPATSPPLLGFNHLTPSQIHQIQSQIQIQQNNH---------------- 916
Query: 241 TSYQNQHSHHH 251
+QNQ+ +HH
Sbjct: 917 LPWQNQNQNHH 949
>TC8599 homologue to UP|Q40478 (Q40478) EREBP-4, partial (26%)
Length = 632
Score = 82.4 bits (202), Expect = 1e-16
Identities = 52/132 (39%), Positives = 74/132 (55%), Gaps = 1/132 (0%)
Frame = +3
Query: 11 PPQEITKPTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKF 70
P I+ P+ + + S S S+ ++ SH ++ ++ ++ DNNK
Sbjct: 243 PSLNISIPSINSGLK---SNISQSKETSFSHQSSSSSSSSDHQTQEE--------DNNK- 386
Query: 71 RYRGVRQRSWGKWVAEIREP-RKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQP 129
YRGVR+R WGK+ AEIR+P RK +R WLGTF TA +AA+AYDRAA + GSRA LN
Sbjct: 387 HYRGVRRRPWGKFAAEIRDPNRKGSRVWLGTFDTAIEAAKAYDRAAFKMRGSRAILNFPL 566
Query: 130 SPTSSSSQSSSS 141
+ + SS
Sbjct: 567 EVGNCHEEEESS 602
>TC13814 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protein, partial
(20%)
Length = 535
Score = 82.0 bits (201), Expect = 1e-16
Identities = 41/73 (56%), Positives = 49/73 (66%)
Frame = +2
Query: 62 KGGPDNNKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGS 121
K G K YRG+RQR WGKW AEIR+PRK R LGTF+TAE+AA+AYD AA + G
Sbjct: 308 KKGKRVRKNLYRGIRQRPWGKWAAEIRDPRKGVRVGLGTFNTAEEAAQAYDDAAKRIRGD 487
Query: 122 RAQLNLQPSPTSS 134
+A+LN P S
Sbjct: 488 KAKLNFPDKPEPS 526
>TC10014 similar to UP|Q8LLR3 (Q8LLR3) Ethylene-responsive element binding
protein 1, partial (33%)
Length = 580
Score = 81.6 bits (200), Expect = 2e-16
Identities = 47/131 (35%), Positives = 74/131 (55%), Gaps = 1/131 (0%)
Frame = +3
Query: 28 PSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRYRGVRQRSWGKWVAEI 87
P + +S + +S NN ++P+ ++ + + P R+RGVR+R WGK+ AEI
Sbjct: 168 PIQDFDSSDGSSQDTNN-SSPSGCKKGSSAEARSSHAPPAEWKRFRGVRRRPWGKFAAEI 344
Query: 88 REPRKR-TRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSPTSSSSQSSSSSSRAS 146
R+P+K R WLGT+ T E+A AYDRAA + G +A+LN P S +Q ++S +
Sbjct: 345 RDPKKNGARVWLGTYETEEEAGLAYDRAAFKIRGRKAKLNF-PHLIGSDTQPETASVVEA 521
Query: 147 SSSSSTQTLRP 157
+S + L P
Sbjct: 522 FKQNSPEPLSP 554
>BP036840
Length = 584
Score = 81.6 bits (200), Expect = 2e-16
Identities = 44/97 (45%), Positives = 61/97 (62%), Gaps = 14/97 (14%)
Frame = +3
Query: 72 YRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQ--- 128
YRGVR+R WG+WV+EIREPRK++R WLG+F E AA+AYD AA L G +AQLN
Sbjct: 87 YRGVRKRRWGRWVSEIREPRKKSRIWLGSFPVPEMAAKAYDVAAYCLKGRKAQLNFPDEV 266
Query: 129 ---PSPT--------SSSSQSSSSSSRASSSSSSTQT 154
P P +++++++S S+A+ SS T
Sbjct: 267 DRLPVPATCTARDIQAAAAKAASMMSKAAGEKSSIAT 377
>TC9504 similar to UP|Q9SXS8 (Q9SXS8) Ethylene responsive element binding
factor, partial (35%)
Length = 633
Score = 80.1 bits (196), Expect = 5e-16
Identities = 37/61 (60%), Positives = 46/61 (74%)
Frame = +2
Query: 71 RYRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPS 130
RYRGVR+R WG++ AEIR+P K+ R WLGTF +AE+AARAYD AA L G +A+ N S
Sbjct: 431 RYRGVRKRPWGRFAAEIRDPMKKARVWLGTFDSAEEAARAYDAAARALRGPKAKTNFPLS 610
Query: 131 P 131
P
Sbjct: 611 P 613
>TC15842 homologue to GB|BAB62912.1|15207792|AB055883 ERF domain protein12
{Arabidopsis thaliana;} , partial (41%)
Length = 746
Score = 79.7 bits (195), Expect = 6e-16
Identities = 36/63 (57%), Positives = 48/63 (76%)
Frame = +2
Query: 72 YRGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNLQPSP 131
YRGVR+R WG++ AEIR+P K+TR WLGTF T E+AA AYD AA L G++A+ N P+P
Sbjct: 185 YRGVRKRPWGRYAAEIRDPWKKTRVWLGTFDTPEEAALAYDGAARSLRGAKAKTNFPPAP 364
Query: 132 TSS 134
++
Sbjct: 365 LAA 373
>TC9975 similar to UP|AAR15448 (AAR15448) AP2 transcription factor, partial
(41%)
Length = 865
Score = 78.2 bits (191), Expect = 2e-15
Identities = 43/88 (48%), Positives = 54/88 (60%), Gaps = 8/88 (9%)
Frame = +1
Query: 71 RYRGVRQRSWGKWVAEIREP-RKRTRKWLGTFSTAEDAARAYDRAAIILYGSRAQLNL-- 127
RYRGVR+R WGK+ AEIR+P RK TR WLGTF T DAA+AYD AA + G +A LN
Sbjct: 181 RYRGVRRRPWGKFAAEIRDPTRKGTRVWLGTFDTEIDAAKAYDYAAFRMRGRKAILNFPL 360
Query: 128 -----QPSPTSSSSQSSSSSSRASSSSS 150
P P++S + + +SSS
Sbjct: 361 EAGKASPKPSNSGRKRGREQRESGTSSS 444
>AV413120
Length = 330
Score = 75.5 bits (184), Expect = 1e-14
Identities = 40/98 (40%), Positives = 51/98 (51%)
Frame = +1
Query: 13 QEITKPTATAAITTTPSETSNSENSNSSHNNNITTPNTSTTINNRKCKGKGGPDNNKFRY 72
Q++T P PS+ S ++S+S+ T T+ T + + RY
Sbjct: 52 QDVTAPNRNIGDFRVPSQQSQPQSSSSA-----TEAATTATSSEASASPSEESGERRRRY 216
Query: 73 RGVRQRSWGKWVAEIREPRKRTRKWLGTFSTAEDAARA 110
RGVRQR WGKW AEIR+P K R WLGT T E AARA
Sbjct: 217 RGVRQRPWGKWAAEIRDPHKAARVWLGTXXTXEAAARA 330
Score = 27.3 bits (59), Expect = 3.8
Identities = 11/33 (33%), Positives = 17/33 (51%)
Frame = +2
Query: 247 HSHHHQDQVVMQNYPVLNHQQHNQNCMGEGVSD 279
H+HH Q Q +Q ++ HQ H+ + G D
Sbjct: 116 HNHHPQQQKQLQQLHLVKHQHHHLKKVVRGGGD 214
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.127 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,979,893
Number of Sequences: 28460
Number of extensions: 160092
Number of successful extensions: 5296
Number of sequences better than 10.0: 429
Number of HSP's better than 10.0 without gapping: 2512
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3776
length of query: 340
length of database: 4,897,600
effective HSP length: 91
effective length of query: 249
effective length of database: 2,307,740
effective search space: 574627260
effective search space used: 574627260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0002.14


