

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588c.4
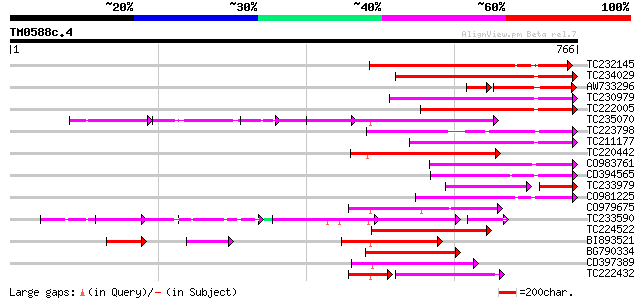
(766 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232145 236 4e-62
TC234029 similar to UP|Q9LN01 (Q9LN01) T6D22.15, partial (32%) 226 2e-59
AW733296 181 6e-57
TC230979 weakly similar to UP|Q6Z8F8 (Q6Z8F8) Selenium-binding p... 217 2e-56
TC222005 weakly similar to UP|Q9FIB2 (Q9FIB2) Selenium-binding p... 198 6e-51
TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 183 2e-46
TC223798 similar to UP|Q9ZQE5 (Q9ZQE5) Expressed protein (At2g15... 181 1e-45
TC211177 weakly similar to UP|Q9LW63 (Q9LW63) Emb|CAB37460.1, pa... 179 4e-45
TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 167 2e-41
CO983761 166 5e-41
CD394565 159 4e-39
TC233979 97 3e-38
CO981225 154 1e-37
CO979675 151 9e-37
TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-... 138 2e-36
TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, pa... 147 1e-35
BI893521 138 8e-33
BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product... 138 1e-32
CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thalian... 133 3e-31
TC222432 92 2e-30
>TC232145
Length = 817
Score = 236 bits (601), Expect = 4e-62
Identities = 120/274 (43%), Positives = 169/274 (60%)
Frame = +3
Query: 487 VHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEA 546
V P+ TFV++LSACSH G +++G + FNSM++ H I P L+HYACMVDL GRVG++ EA
Sbjct: 9 VVPEYITFVSVLSACSHTGKIDDGFKYFNSMANVHNIKPGLEHYACMVDLLGRVGRLEEA 188
Query: 547 EDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSE 606
I +MP +PDS++W +LLG+C KH + R AE+ +L+P N Y+ +SNIY
Sbjct: 189 CRFIESMPFEPDSLVWGALLGACGKHANVEMGREVAERLFKLEPDNPGNYMLLSNIYIRH 368
Query: 607 GSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQL 666
G EA +R+ M + V+K+ G SW++V + F + H + I L+ L +
Sbjct: 369 GMLEEADEVRRLMGINGVRKESGCSWIDVKNRTFVFNANDRSHSRTQEIYGMLQKLKELI 548
Query: 667 KEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRIC 726
K GYV E A E +E L+ HSEK+AL F ++ LP G+ ++I KN+R C
Sbjct: 549 KRRGYVAETQFATNSVE-GSEEQSLWCHSEKLALAFGLL---VLP-PGSPVRIKKNLRTC 713
Query: 727 ADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATC 760
DCH MK AS +FQ+EI+VRD NRFH F N +C
Sbjct: 714 GDCHTVMKFASEIFQREIIVRDINRFHRFTNGSC 815
Score = 39.3 bits (90), Expect = 0.007
Identities = 53/226 (23%), Positives = 84/226 (36%), Gaps = 15/226 (6%)
Frame = +3
Query: 387 ENFVPDWHTFSIALKACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLAL 446
E VP++ TF L AC+ H+ I GF+ ++N +H
Sbjct: 3 EGVVPEYITFVSVLSACS-----------HTGKIDDGFKYFNSMAN--VHNIKPG----- 128
Query: 447 SEQVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKLDVHPDSTTFVALLSACSHAGL 506
L + M+ G+ ++A + + PDS + ALL AC
Sbjct: 129 ------------LEHYACMVDLLGRVGRLEEACRFIESMPFEPDSLVWGALLGACGKHAN 272
Query: 507 VEEGVEI----FNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPM----KPD 558
VE G E+ F DN G +Y + ++Y R G + EA+++ M + K
Sbjct: 273 VEMGREVAERLFKLEPDNPG------NYMLLSNIYIRHGMLEEADEVRRLMGINGVRKES 434
Query: 559 SVIWSSLL-------GSCRKHGETRLARIAAEKFKELDPKNSLGYV 597
W + + R H T+ +K KEL + GYV
Sbjct: 435 GCSWIDVKNRTFVFNANDRSHSRTQEIYGMLQKLKELIKRR--GYV 566
>TC234029 similar to UP|Q9LN01 (Q9LN01) T6D22.15, partial (32%)
Length = 732
Score = 226 bits (577), Expect = 2e-59
Identities = 109/245 (44%), Positives = 152/245 (61%)
Frame = +2
Query: 522 GIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIA 581
GI P+L HY CM+DL R GK EA+ L+ M M+PD IW SLL +CR HG+
Sbjct: 2 GISPKLQHYGCMIDLLARSGKFDEAKVLMGNMEMEPDGAIWGSLLNACRIHGQVEFGEYV 181
Query: 582 AEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHE 641
AE+ EL+P+NS YV +SNIY+ G + + IR ++ D +KK PG + +E+ VHE
Sbjct: 182 AERLFELEPENSGAYVLLSNIYAGAGRWDDVAKIRTKLNDKGMKKVPGCTSIEIDGVVHE 361
Query: 642 FTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALV 701
F G HP E I L+ + L+E G+VP+ + LYD + E KE L HSEK+A+
Sbjct: 362 FLVGDKFHPQSENIFRMLDEVDRLLEETGFVPDTSEVLYDMDEEWKEGALTQHSEKLAIA 541
Query: 702 FAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCS 761
F +++ G+ I+I+KN+R+C +CH+ KL S +F +EI+ RD NRFHHFK+ CS
Sbjct: 542 FGLISTK----PGSTIRIVKNLRVCRNCHSATKLISKIFNREIIARDRNRFHHFKDGFCS 709
Query: 762 CNDYW 766
CND W
Sbjct: 710 CNDCW 724
Score = 31.2 bits (69), Expect = 1.8
Identities = 23/88 (26%), Positives = 38/88 (43%), Gaps = 4/88 (4%)
Frame = +2
Query: 459 LVSWNSMLKSYALHGKAKDALELFKKLDVHPDSTTFVALLSACSHAGLVEEG----VEIF 514
L + M+ A GK +A L +++ PD + +LL+AC G VE G +F
Sbjct: 17 LQHYGCMIDLLARSGKFDEAKVLMGNMEMEPDGAIWGSLLNACRIHGQVEFGEYVAERLF 196
Query: 515 NSMSDNHGIVPQLDHYACMVDLYGRVGK 542
+N G Y + ++Y G+
Sbjct: 197 ELEPENSGA------YVLLSNIYAGAGR 262
>AW733296
Length = 424
Score = 181 bits (458), Expect(2) = 6e-57
Identities = 90/112 (80%), Positives = 95/112 (84%)
Frame = +3
Query: 654 AIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCG 713
A+ SRLEI+IGQLKEMGYVPE++LALYDTEVEHKEDQL HHS+KMALVFAIMNEG
Sbjct: 111 AMLSRLEIVIGQLKEMGYVPELSLALYDTEVEHKEDQLLHHSKKMALVFAIMNEG----- 275
Query: 714 GNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDY 765
IKIMKNIRIC DCHNFMKLAS LFQKEI RDSN FHHFK A CSCNDY
Sbjct: 276 ---IKIMKNIRICVDCHNFMKLASYLFQKEIAARDSNCFHHFKYAACSCNDY 422
Score = 59.7 bits (143), Expect(2) = 6e-57
Identities = 24/34 (70%), Positives = 30/34 (87%)
Frame = +2
Query: 618 EMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPH 651
EM D +V+K+PGLSWVE+GKQVHEF SGG +HP+
Sbjct: 5 EMSDYKVRKEPGLSWVEIGKQVHEFGSGGQYHPN 106
>TC230979 weakly similar to UP|Q6Z8F8 (Q6Z8F8) Selenium-binding protein-like,
partial (61%)
Length = 837
Score = 217 bits (552), Expect = 2e-56
Identities = 102/253 (40%), Positives = 151/253 (59%)
Frame = +2
Query: 514 FNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHG 573
FN M H I P + HY CMVDL GR G + EA DLI +MP+KP+ V+W SLL +C+ H
Sbjct: 5 FNRMQFEHMIKPTIQHYGCMVDLMGRAGMLKEAYDLIKSMPIKPNDVVWRSLLSACKVHH 184
Query: 574 ETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWV 633
+ IAA+ +L+ N Y+ ++N+Y+ + IR EM + + + PG S V
Sbjct: 185 NLEIGEIAADNIFKLNKHNPGDYLVLANMYARAQKWANVARIRTEMVEKNLVQTPGFSLV 364
Query: 634 EVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFH 693
E + V++F S P E I ++ + QLK GY P+++ L D + + K +L H
Sbjct: 365 EANRNVYKFVSQDKSQPKCETIYDMIQQMEWQLKFEGYTPDMSQVLLDVDEDEKRQRLKH 544
Query: 694 HSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFH 753
HS+K+A+ FA++ G+ I+I +N+R+C DCH + K S ++++EI VRD NRFH
Sbjct: 545 HSQKLAIAFALIQTSE----GSPIRISRNLRMCNDCHTYTKFISVIYEREITVRDRNRFH 712
Query: 754 HFKNATCSCNDYW 766
HFK+ TCSC DYW
Sbjct: 713 HFKDGTCSCKDYW 751
Score = 33.1 bits (74), Expect = 0.47
Identities = 16/61 (26%), Positives = 32/61 (52%)
Frame = +2
Query: 128 YARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAHYRPNEFAFASLLSACEE 187
+ R F+ M + I + ++ + G+++E + L + +PN+ + SLLSAC+
Sbjct: 5 FNRMQFEHMIKPTIQHYGCMVDLMGRAGMLKEAYDLIKSM--PIKPNDVVWRSLLSACKV 178
Query: 188 H 188
H
Sbjct: 179 H 181
>TC222005 weakly similar to UP|Q9FIB2 (Q9FIB2) Selenium-binding protein-like,
partial (13%)
Length = 757
Score = 198 bits (504), Expect = 6e-51
Identities = 93/212 (43%), Positives = 133/212 (61%)
Frame = +2
Query: 555 MKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGL 614
+KPD +W+SLLG CR HG LA+ AA+ E++P+N Y+ ++NIY++ G + E
Sbjct: 8 VKPDKFLWASLLGGCRIHGNLELAKRAAKALYEIEPENPATYITLANIYANAGLWSEVAN 187
Query: 615 IRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPE 674
+RK+M + + K+PG SW+E+ +QVH F G HP I L L ++KE GYVP+
Sbjct: 188 VRKDMDNMGIVKKPGKSWIEIKRQVHVFLVGDTSHPKTSDIHEFLGELSKKIKEEGYVPD 367
Query: 675 ITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMK 734
L+D E E KE L +HSEK+A+ F I++ G IK+ KN+R C DCH +K
Sbjct: 368 TNFVLHDVEEEQKEQNLVYHSEKLAVAFGIISTP----PGTPIKVFKNLRTCVDCHTAIK 535
Query: 735 LASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
S + Q++I VRDSNRFH F++ +CSC DYW
Sbjct: 536 YISKIVQRKITVRDSNRFHCFEDGSCSCKDYW 631
>TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (22%)
Length = 875
Score = 183 bits (465), Expect = 2e-46
Identities = 89/262 (33%), Positives = 155/262 (58%), Gaps = 3/262 (1%)
Frame = +2
Query: 402 ACAYFVTEQQALAVHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCCHDLVS 461
AC+ + +Q +H+ +IK FQ + + AL+ Y++ G LA +++ F + ++ +
Sbjct: 2 ACSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAA 181
Query: 462 WNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIFNSMS 518
W +++ YA HG +A+ LF+ + + P++ TFV +LSAC+HAGLV EG+ IF+SM
Sbjct: 182 WTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSMQ 361
Query: 519 DNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLA 578
+G+ P ++HY C+VDL GR G + EAE+ I MP++ D +IW +LL + + +
Sbjct: 362 RCYGVTPTIEHYTCVVDLLGRSGHLKEAEEFIIKMPIEADGIIWGALLNASWFWKDMEVG 541
Query: 579 RIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQ 638
AAEK LDP +V +SN+Y+ G + + +RK ++ ++K PG SW+E+ +
Sbjct: 542 ERAAEKLFSLDPNPIFAFVVLSNMYAILGRWGQKTKLRKRLQSLELRKDPGCSWIELNNK 721
Query: 639 VHEFTSGGHHHPHKEAIQSRLE 660
+H F+ H + + I + +E
Sbjct: 722 IHLFSVEDKTHLYSDVIYATVE 787
Score = 70.5 bits (171), Expect = 3e-12
Identities = 43/112 (38%), Positives = 61/112 (54%), Gaps = 1/112 (0%)
Frame = +2
Query: 82 ACAKNKCLQQGMALHNYILHKDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQMPRRNI 141
AC+ +QG LH +++ K P Q ++++ LV+ YSKCGHL A+ F + N+
Sbjct: 2 ACSCLCSFRQGQLLHAHLI-KTP-FQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNV 175
Query: 142 VSWTALISGYAQCGLIRECFYLFSGLLAH-YRPNEFAFASLLSACEEHDIKC 192
+WTALI+GYA GL E LF +L PN F +LSAC + C
Sbjct: 176 AAWTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVC 331
Score = 61.2 bits (147), Expect = 2e-09
Identities = 46/174 (26%), Positives = 74/174 (42%)
Frame = +2
Query: 193 GMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTGDDAWTMFKSMEFRNLISW 252
G +HA +K +VYV AL+ YSKC H +A F S+ N+ +W
Sbjct: 32 GQLLHAHLIKTPFQVNVYVGTALVDFYSKCG---HLA------EAQRSFISIFSPNVAAW 184
Query: 253 NSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSAFDELNIPLRNCFV 312
++I G+ + GLG +AI LF M GI + AT + V S+ N E +
Sbjct: 185 TALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCE------GLRI 346
Query: 313 LHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAII 366
H + G+ +E T +V G + + + + D + W A++
Sbjct: 347 FHSMQRCYGVTPTIEHYTCVV-DLLGRSGHLKEAEEFIIKMPIEADGIIWGALL 505
Score = 43.1 bits (100), Expect = 5e-04
Identities = 43/160 (26%), Positives = 69/160 (42%), Gaps = 3/160 (1%)
Frame = +2
Query: 312 VLHCLATKTGLISEVEVVTALVKSYANLGGQISDCYRLFLDTSGKQDIVSWTAIITVLAD 371
+LH KT V V TALV Y+ G +++ R F+ ++ +WTA+I A
Sbjct: 38 LLHAHLIKTPFQVNVYVGTALVDFYSKCG-HLAEAQRSFISIFSP-NVAAWTALINGYAY 211
Query: 372 QD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALAV-HSQVIKRGFQKDTV 429
+A LLF + + VP+ TF L AC + + L + HS G
Sbjct: 212 HGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSMQRCYGVTPTIE 391
Query: 430 LSNALIHAYARSGSLALSEQVFDEMCCH-DLVSWNSMLKS 468
++ RSG L +E+ +M D + W ++L +
Sbjct: 392 HYTCVVDLLGRSGHLKEAEEFIIKMPIEADGIIWGALLNA 511
>TC223798 similar to UP|Q9ZQE5 (Q9ZQE5) Expressed protein
(At2g15690/F9O13.24), partial (16%)
Length = 789
Score = 181 bits (459), Expect = 1e-45
Identities = 98/284 (34%), Positives = 156/284 (54%)
Frame = +3
Query: 483 KKLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGK 542
K+ V PD TF +L+AC+ A VEEG F SM + HGIVP ++HY ++++ G G+
Sbjct: 9 KQAGVPPDGETFELVLAACAQAEAVEEGFLHFESMKE-HGIVPSMEHYLEVINILGNAGQ 185
Query: 543 ISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNI 602
++EAE+ I +P++ W SL +KHG+ L A E LDP +
Sbjct: 186 LNEAEEFIEKIPIELGVEAWESLRNFAQKHGDLDLEDHAEEVLTCLDPSKA--------- 338
Query: 603 YSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEIL 662
+ ++ KKQ ++ +E +V E+ + P+KE +L L
Sbjct: 339 ------------VADKLPPPPRKKQSDVNMLEEKNRVTEYR---YSIPYKEEAHEKLGGL 473
Query: 663 IGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKN 722
GQ++E GYVP+ L+D + + KE L +HSE++A+ + +++ ++I+KN
Sbjct: 474 SGQMREAGYVPDTRYVLHDIDEDDKEKALQYHSERLAIAYGLISTPPR----TTLRIIKN 641
Query: 723 IRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
+RIC DCHN +K+ S + +E++VRD+ RFHHFK+ CSC DYW
Sbjct: 642 LRICGDCHNAIKIMSKIVGRELIVRDNKRFHHFKDGKCSCGDYW 773
>TC211177 weakly similar to UP|Q9LW63 (Q9LW63) Emb|CAB37460.1, partial (17%)
Length = 823
Score = 179 bits (454), Expect = 4e-45
Identities = 89/227 (39%), Positives = 130/227 (57%), Gaps = 1/227 (0%)
Frame = +2
Query: 541 GKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMS 600
G++ EA I +P+KP W +LL SC HG +A++ ++ ELD + YV +S
Sbjct: 5 GRLEEACKFIDELPIKPTPXXWRTLLSSCSSHGNVEMAKLVIQRIFELDDSHGGDYVILS 184
Query: 601 NIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLE 660
N+ + G + + +RK M D K PG S +EV VHEF SG H + L+
Sbjct: 185 NLCARNGRWDDVNHLRKMMVDKGALKVPGCSSIEVNNVVHEFFSGDGVHSTSTILHHALD 364
Query: 661 ILIGQLKEMGYVPEITLALY-DTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKI 719
L+ +LK GYVP+ +L Y D E E KE L +HSEK+A+ + ++N G I++
Sbjct: 365 ELVKELKLAGYVPDTSLVFYADIEDEEKEIVLRYHSEKLAITYGLLNTP----PGTTIRV 532
Query: 720 MKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
+KN+R+C DCHN K S +F ++I++RD RFHH K+ CSC DYW
Sbjct: 533 VKNLRVCVDCHNAAKFISLIFGRQIILRDVQRFHHLKDGKCSCGDYW 673
Score = 30.0 bits (66), Expect = 4.0
Identities = 20/80 (25%), Positives = 36/80 (45%), Gaps = 3/80 (3%)
Frame = +2
Query: 473 GKAKDALELFKKLDVHPDSTTFVALLSACSHAGLVEEG---VEIFNSMSDNHGIVPQLDH 529
G+ ++A + +L + P + LLS+CS G VE ++ + D+HG
Sbjct: 5 GRLEEACKFIDELPIKPTPXXWRTLLSSCSSHGNVEMAKLVIQRIFELDDSHG-----GD 169
Query: 530 YACMVDLYGRVGKISEAEDL 549
Y + +L R G+ + L
Sbjct: 170 YVILSNLCARNGRWDDVNHL 229
>TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (25%)
Length = 706
Score = 167 bits (422), Expect = 2e-41
Identities = 78/205 (38%), Positives = 126/205 (61%), Gaps = 3/205 (1%)
Frame = +1
Query: 461 SWNSMLKSYALHGKAKDALELF---KKLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSM 517
+W +++ A+HGK ++AL+ F +K ++P+S TF A+L+ACSHAGL EEG +F SM
Sbjct: 16 AWTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSHAGLTEEGKSLFESM 195
Query: 518 SDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRL 577
S + I P ++HY CMVDL GR G + EA + I +MP+KP++ IW +LL +C+ H L
Sbjct: 196 SSVYNIKPSMEHYGCMVDLMGRAGLLKEAREFIESMPVKPNAAIWGALLNACQLHKHFEL 375
Query: 578 ARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGK 637
+ + ELDP +S Y+ +++IY++ G + + +R +++ + PG S + +
Sbjct: 376 GKEIGKILIELDPDHSGRYIHLASIYAAAGEWNQVVRVRSQIKHRGLLNHPGCSSITLNG 555
Query: 638 QVHEFTSGGHHHPHKEAIQSRLEIL 662
VHEF +G HPH + I +L
Sbjct: 556 VVHEFFAGDGSHPHIQEIYGMPNLL 630
Score = 32.3 bits (72), Expect = 0.80
Identities = 17/49 (34%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Frame = +1
Query: 138 RRNIVSWTALISGYAQCGLIRECFYLFSGLL-AHYRPNEFAFASLLSAC 185
++ + +WTA+I G A G RE F+ + A PN F ++L+AC
Sbjct: 1 KKCVCAWTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTAC 147
>CO983761
Length = 775
Score = 166 bits (419), Expect = 5e-41
Identities = 82/199 (41%), Positives = 117/199 (58%)
Frame = -2
Query: 568 SCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQ 627
SCR H LA A+ EL+P+N+ YV +++IY+ + EA + K + ++K
Sbjct: 774 SCRIHCNVELAERASTLLFELEPRNAGNYVLLADIYAEAKMWSEAKSVMKLLEARGLQKL 595
Query: 628 PGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHK 687
PG SW+EV ++V+ F S H+P E I + L L ++K GYVP+ + LYD + E K
Sbjct: 594 PGCSWIEVKRKVYSFVSVDEHNPQIEEIHALLVKLSNEMKAQGYVPQTNVVLYDLDEEEK 415
Query: 688 EDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVR 747
E + HSEK+A+ F ++N G I+I KN+R+C DCH K S +EI+VR
Sbjct: 414 ERIVLGHSEKLAVAFGLIN----TVKGETIRIRKNLRLCEDCHAVTKFISKFANREILVR 247
Query: 748 DSNRFHHFKNATCSCNDYW 766
D NRFHHFK+ CSC DYW
Sbjct: 246 DVNRFHHFKDGVCSCGDYW 190
>CD394565
Length = 682
Score = 159 bits (402), Expect = 4e-39
Identities = 78/198 (39%), Positives = 115/198 (57%)
Frame = -3
Query: 569 CRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQP 628
C+ H L AA K +LDP +V ++NIY+S + + +R M D + K P
Sbjct: 680 CKIHKNVELGEKAAHKLFKLDPDEGGYHVLLANIYASNSMWDKVAKVRTAMEDKGLHKTP 501
Query: 629 GLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKE 688
G SWVE+ ++H F SG +HP + I + LE L ++K GYVP+ +++D E + K+
Sbjct: 500 GCSWVELRNEIHTFYSGSTNHPESKKIYAFLETLGDEIKAAGYVPDPD-SIHDVEEDVKK 324
Query: 689 DQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRICADCHNFMKLASNLFQKEIVVRD 748
L HSE++A+ F ++N G + I KN+R+C DCH+ K S + +EI+VRD
Sbjct: 323 QLLSSHSERLAIAFGLLNTS----PGTTLHIRKNLRVCGDCHDTTKYISLVTGREIIVRD 156
Query: 749 SNRFHHFKNATCSCNDYW 766
RFHHFKN +CSC DYW
Sbjct: 155 LRRFHHFKNGSCSCGDYW 102
>TC233979
Length = 825
Score = 97.4 bits (241), Expect(2) = 3e-38
Identities = 43/117 (36%), Positives = 70/117 (59%)
Frame = +3
Query: 589 DPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHH 648
+P +S YV +SN+Y++ + R MR + VKK PG SWV +VH F +G
Sbjct: 27 EPSDSAAYVLLSNVYAAANQWENVASARNMMRKANVKKDPGFSWVXXKNKVHLFVAGDRS 206
Query: 649 HPHKEAIQSRLEILIGQLKEMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIM 705
H + I +++E ++ +++E GY+P+ AL D E E KE L++HSEK+A+ + +M
Sbjct: 207 HEETDVIYNKVEYIMKRIREEGYLPDTDFALVDVEEEDKECSLYYHSEKLAIAYGLM 377
Score = 80.5 bits (197), Expect(2) = 3e-38
Identities = 27/50 (54%), Positives = 42/50 (84%)
Frame = +1
Query: 717 IKIMKNIRICADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
++++KN+R+C DCHN +K S +F++E+V+RD+NRFHHF++ CSC DYW
Sbjct: 400 LRVIKNLRVCGDCHNAIKYISKVFEREVVLRDANRFHHFRSGVCSCGDYW 549
>CO981225
Length = 754
Score = 154 bits (390), Expect = 1e-37
Identities = 84/220 (38%), Positives = 116/220 (52%), Gaps = 2/220 (0%)
Frame = -2
Query: 549 LIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGS 608
+I +MP +PD + W SLLG C + IAA+ LDP +S YV M N+Y+ G
Sbjct: 753 VIRSMPFEPDVMSWKSLLGGCWSRRNLEIGMIAADNIFRLDPLDSATYVIMFNLYALAGK 574
Query: 609 FIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLK- 667
+ EA RK M + ++K+ SW+ V +VH F G HHP E I S+L+ L K
Sbjct: 573 WDEAAQFRKMMAERNLRKEVSCSWIIVKGKVHRFVVGDRHHPQTEQIYSKLKELNVSFKK 394
Query: 668 -EMGYVPEITLALYDTEVEHKEDQLFHHSEKMALVFAIMNEGNLPCGGNVIKIMKNIRIC 726
E + E TE ++DQL HSE++A+ + ++ I + KN R C
Sbjct: 393 GEERLLNEENALCDFTE---RKDQLLDHSERLAIAYGLI----CTAADTPIMVFKNTRSC 235
Query: 727 ADCHNFMKLASNLFQKEIVVRDSNRFHHFKNATCSCNDYW 766
DCH F K S + +E+VVRD NRFHH + CSC DYW
Sbjct: 234 KDCHEFAKRVSVVTGRELVVRDGNRFHHINSGECSCRDYW 115
>CO979675
Length = 732
Score = 151 bits (382), Expect = 9e-37
Identities = 79/217 (36%), Positives = 126/217 (57%), Gaps = 8/217 (3%)
Frame = -2
Query: 458 DLVSWNSMLKSYALHGKAKDALELFKKLD---VHPDSTTFVALLSACSHAGLVEEGVEIF 514
D+V WN+M+ A+HG AL++F +++ + PD TF+A+ +ACS++G+ EG+++
Sbjct: 728 DIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQLL 549
Query: 515 NSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPM-----KPDSVIWSSLLGSC 569
+ MS + I P+ +HY C+VDL R G EA +I + +++ W + L +C
Sbjct: 548 DKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAFLSAC 369
Query: 570 RKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPG 629
HG+ +LA AA++ L+ + + YV +SN+Y++ G +A +R MR+ V K PG
Sbjct: 368 CNHGQAQLAERAAKRLLRLENHSGV-YVLLSNLYAASGKHSDARRVRNMMRNKGVDKAPG 192
Query: 630 LSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQL 666
S VE+ V EF +G HP E I S LEIL QL
Sbjct: 191 CSSVEIDGVVSEFIAGEETHPQMEEIHSVLEILHMQL 81
Score = 30.4 bits (67), Expect = 3.1
Identities = 16/48 (33%), Positives = 26/48 (53%), Gaps = 1/48 (2%)
Frame = -2
Query: 139 RNIVSWTALISGYAQCGLIRECFYLFSGL-LAHYRPNEFAFASLLSAC 185
R+IV W A+ISG A G +FS + +P++ F ++ +AC
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTAC 588
>TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-containing
protein-like, partial (5%)
Length = 1094
Score = 138 bits (348), Expect(2) = 2e-36
Identities = 79/259 (30%), Positives = 140/259 (53%), Gaps = 5/259 (1%)
Frame = +2
Query: 356 KQDIVSWTAIITVLADQDPE-QAFLLFCQLHRENFVPDWHTFSIALKACAYFVTEQQALA 414
K D SWT++I A +A +F Q+ + PD T S L ACA + +
Sbjct: 65 KSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQFTLSTCLFACATIASLKHGRQ 244
Query: 415 VHSQVIKRGFQKDTVLSNALIHAYARSGSLALSEQVFDEMCC-HDLVSWNSMLKSYALHG 473
+H+ ++ + +T++ A+++ Y++ GSL + +VF+ + D+V WN+M+ + A +G
Sbjct: 245 IHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFNFIGNKQDVVLWNTMILALAHYG 424
Query: 474 KAKDALELFK---KLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHY 530
+A+ + K+ V P+ TFV +L+AC H+GLV+EG+++F SM+ HG+VP +HY
Sbjct: 425 YGIEAIMMLYNMLKIGVKPNKGTFVGILNACCHSGLVQEGLQLFKSMTSEHGVVPDQEHY 604
Query: 531 ACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDP 590
+ +L G+ +E+ + M KP + +S +G CR HG A +L P
Sbjct: 605 TRLANLLGQARCFNESVKDLQMMDCKPGDHVCNSSIGVCRMHGNIDHGAEVAAFLIKLQP 784
Query: 591 KNSLGYVQMSNIYSSEGSF 609
++S Y +S Y++ G +
Sbjct: 785 QSSAAYELLSRTYAALGKW 841
Score = 83.6 bits (205), Expect = 3e-16
Identities = 64/228 (28%), Positives = 112/228 (49%), Gaps = 2/228 (0%)
Frame = +2
Query: 117 VNMYSKCGHLEYARYVFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLLAH-YRPNE 175
V+ Y+ G +E +F QMP+ + SWT+LI GYA+ G+ E +F ++ H RP++
Sbjct: 2 VSGYAVWGDMESGAELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQ 181
Query: 176 FAFASLLSACEE-HDIKCGMQVHAVALKISLDASVYVANALITMYSKCSGGFHGGYDPTG 234
F ++ L AC +K G Q+HA + ++ + V A++ MYSKC G T
Sbjct: 182 FTLSTCLFACATIASLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKC------GSLETA 343
Query: 235 DDAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSL 294
+ + + +++ WN+MI G G +AI + +M G+ ++ T + + L
Sbjct: 344 RRVFNFIGNKQ--DVVLWNTMILALAHYGYGIEAIMMLYNMLKIGVKPNKGTFVGI---L 508
Query: 295 NECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQ 342
N C + L+ + + ++ G++ + E T L ANL GQ
Sbjct: 509 NACCHSGLVQEGLQ---LFKSMTSEHGVVPDQEHYTRL----ANLLGQ 631
Score = 61.6 bits (148), Expect = 1e-09
Identities = 45/146 (30%), Positives = 72/146 (48%), Gaps = 2/146 (1%)
Frame = +2
Query: 42 IRALSLQGNLEEALSLLYTHDKHILTHSSLSLQTYASLFHACAKNKCLQQGMALHNYILH 101
IR + G EAL + KH + +L T ACA L+ G +H +++
Sbjct: 95 IRGYARNGMGYEALGVFKQMIKHQVRPDQFTLST---CLFACATIASLKHGRQIHAFLVL 265
Query: 102 KDPTIQNDLFLTNHLVNMYSKCGHLEYARYVFDQM-PRRNIVSWTALISGYAQCGL-IRE 159
+ I+ + + +VNMYSKCG LE AR VF+ + ++++V W +I A G I
Sbjct: 266 NN--IKPNTIVVCAIVNMYSKCGSLETARRVFNFIGNKQDVVLWNTMILALAHYGYGIEA 439
Query: 160 CFYLFSGLLAHYRPNEFAFASLLSAC 185
L++ L +PN+ F +L+AC
Sbjct: 440 IMMLYNMLKIGVKPNKGTFVGILNAC 517
Score = 52.8 bits (125), Expect = 6e-07
Identities = 70/284 (24%), Positives = 113/284 (39%), Gaps = 13/284 (4%)
Frame = +2
Query: 229 GYDPTGD--DAWTMFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRAT 286
GY GD +F M + SW S+I G+ G+G +A+ +F M + D+ T
Sbjct: 8 GYAVWGDMESGAELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQFT 187
Query: 287 LLSVFSSLNECSAFDELNIPLRNCFVLHCLATKTGLISEVEVVTALVKSYANLGGQISDC 346
L + L C+ L ++ +H + VV A+V Y+ G +
Sbjct: 188 LSTC---LFACATIASL----KHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGS-LETA 343
Query: 347 YRLFLDTSGKQDIVSWTAIITVLADQD-PEQAFLLFCQLHRENFVPDWHTFSIALKACAY 405
R+F KQD+V W +I LA +A ++ + + P+ TF L AC +
Sbjct: 344 RRVFNFIGNKQDVVLWNTMILALAHYGYGIEAIMMLYNMLKIGVKPNKGTFVGILNACCH 523
Query: 406 FVTEQQALAV-HSQVIKRGFQKD----TVLSNALIHAYARSGSL----ALSEQVFDEMCC 456
Q+ L + S + G D T L+N L A + S+ + + D +C
Sbjct: 524 SGLVQEGLQLFKSMTSEHGVVPDQEHYTRLANLLGQARCFNESVKDLQMMDCKPGDHVC- 700
Query: 457 HDLVSWNSMLKSYALHGKAKDALELFKKL-DVHPDSTTFVALLS 499
NS + +HG E+ L + P S+ LLS
Sbjct: 701 ------NSSIGVCRMHGNIDHGAEVAAFLIKLQPQSSAAYELLS 814
Score = 33.1 bits (74), Expect(2) = 2e-36
Identities = 21/55 (38%), Positives = 29/55 (52%)
Frame = +1
Query: 619 MRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPHKEAIQSRLEILIGQLKEMGYVP 673
MRD K Q +SW+E+ +VH FT HP KE S L L +++ G+ P
Sbjct: 874 MRDRGGKGQ-AISWIEIDNKVHTFTVLDASHPLKETTYSALWALSYRME--GHAP 1029
>TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, partial (18%)
Length = 534
Score = 147 bits (372), Expect = 1e-35
Identities = 69/162 (42%), Positives = 100/162 (61%)
Frame = +2
Query: 490 DSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDL 549
D TF++LLSAC AG V E + +F+ M DN+GI P+ +HYAC+VD+ R G++ A +
Sbjct: 2 DGITFLSLLSACCRAGKVNESMNLFSLMVDNYGIPPRSEHYACLVDVMSRAGQLQRACKI 181
Query: 550 IHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMSNIYSSEGSF 609
I+ MP K DS IW ++L +C H L +AA + LDP NS YV +SNIY++ G +
Sbjct: 182 INEMPFKADSSIWGAVLAACSVHLNVELGELAARRILNLDPFNSGAYVMLSNIYAAAGKW 361
Query: 610 IEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHEFTSGGHHHPH 651
+ IR M++ VKKQ SW+++G + H F G HP+
Sbjct: 362 KDVHRIRVLMKEQGVKKQTAYSWLQIGNKTHYFVGGDPSHPN 487
>BI893521
Length = 423
Score = 138 bits (348), Expect = 8e-33
Identities = 65/139 (46%), Positives = 96/139 (68%), Gaps = 3/139 (2%)
Frame = +1
Query: 449 QVFDEMCCHDLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAG 505
QVF++M ++++W S++ +A HG A ALELF ++ V P+ T++A+LSACSH G
Sbjct: 1 QVFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVG 180
Query: 506 LVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSL 565
L++E + FNSM NH I P+++HYACMVDL GR G + EA + I++MP D+++W +
Sbjct: 181 LIDEAWKHFNSMHYNHSISPRMEHYACMVDLLGRSGLLLEAIEFINSMPFDADALVWRTF 360
Query: 566 LGSCRKHGETRLARIAAEK 584
LGSCR H T+L AA+K
Sbjct: 361 LGSCRVHRNTKLGEHAAKK 417
Score = 47.4 bits (111), Expect = 2e-05
Identities = 22/55 (40%), Positives = 36/55 (65%), Gaps = 1/55 (1%)
Frame = +1
Query: 132 VFDQMPRRNIVSWTALISGYAQCGLIRECFYLFSGLL-AHYRPNEFAFASLLSAC 185
VF+ M RN+++WT++ISG+A+ G + LF +L +PNE + ++LSAC
Sbjct: 4 VFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSAC 168
Score = 47.0 bits (110), Expect = 3e-05
Identities = 22/63 (34%), Positives = 35/63 (54%)
Frame = +1
Query: 240 MFKSMEFRNLISWNSMIAGFQFRGLGDKAIRLFAHMYCSGIGFDRATLLSVFSSLNECSA 299
+F M +RN+I+W S+I+GF G KA+ LF M G+ + T ++V S+ +
Sbjct: 4 VFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVGL 183
Query: 300 FDE 302
DE
Sbjct: 184 IDE 192
>BG790334 weakly similar to GP|2160154|gb| F19K23.18 gene product
{Arabidopsis thaliana}, partial (19%)
Length = 390
Score = 138 bits (347), Expect = 1e-32
Identities = 61/129 (47%), Positives = 93/129 (71%)
Frame = +2
Query: 481 LFKKLDVHPDSTTFVALLSACSHAGLVEEGVEIFNSMSDNHGIVPQLDHYACMVDLYGRV 540
L K+L +HP TF+++L+AC+HAGLVEEG F SM +++GI P+++H+A +VD+ GR
Sbjct: 2 LMKRLKIHPTYITFISVLNACAHAGLVEEGWRQFKSMINDYGIEPRVEHFASLVDILGRQ 181
Query: 541 GKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIAAEKFKELDPKNSLGYVQMS 600
G++ EA DLI+TMP KPD +W +LLG+CR H LA +AA+ L+P++S YV +
Sbjct: 182 GQLQEAMDLINTMPFKPDKAVWGALLGACRVHNNVELALVAADALIRLEPESSAPYVLLY 361
Query: 601 NIYSSEGSF 609
N+Y++ G +
Sbjct: 362 NMYANLGQW 388
>CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thaliana}, partial
(16%)
Length = 626
Score = 133 bits (334), Expect = 3e-31
Identities = 70/177 (39%), Positives = 106/177 (59%), Gaps = 5/177 (2%)
Frame = -2
Query: 462 WNSMLKSYALHGKAKDALELFKKLDVH----PDSTTFVALLSACSHAGLVEEGVEIFNSM 517
W SM+ Y +G +ALELF K+ P+ T ++ LSAC+HAGLV++G EI SM
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSACAHAGLVDKGWEIIQSM 446
Query: 518 SDNHGIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRL 577
+ + + P ++HYACMVDL GR G +++A + I +P KP S +W++LL SCR HG L
Sbjct: 445 ENEYLVKPGMEHYACMVDLLGRAGMLNQAWEFIMRIPEKPISDVWAALLSSCRLHGNIEL 266
Query: 578 ARIAAEKFKELDPKNSLG-YVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWV 633
A++AA + +L+ G YV +SN + G + +R+ M++ + K SWV
Sbjct: 265 AKLAANELFKLNATGRPGAYVALSNTLVAAG*WESVTELREIMKERGISKDTXRSWV 95
Score = 29.6 bits (65), Expect = 5.2
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 2/44 (4%)
Frame = -2
Query: 144 WTALISGYAQCGLIRECFYLFSGLLAHYR--PNEFAFASLLSAC 185
WT+++ GY + G E LF + Y PN S LSAC
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSAC 494
>TC222432
Length = 951
Score = 91.7 bits (226), Expect(2) = 2e-30
Identities = 48/147 (32%), Positives = 79/147 (53%)
Frame = +2
Query: 522 GIVPQLDHYACMVDLYGRVGKISEAEDLIHTMPMKPDSVIWSSLLGSCRKHGETRLARIA 581
G PQ++HY CMVDL GR G + EAE+LI TMP + +I SS L +C + A
Sbjct: 203 GFAPQVEHYGCMVDLLGRAGCLDEAENLIQTMPYDANGIILSSFLFACGYFNDVLRAERV 382
Query: 582 AEKFKELDPKNSLGYVQMSNIYSSEGSFIEAGLIRKEMRDSRVKKQPGLSWVEVGKQVHE 641
++ ++D + YV + N+Y++ + + +++ M+ K+ S +E+G E
Sbjct: 383 LKEVVKMDEDVAGNYVMLRNLYATRQRWTDVEDVKQMMKKRGTSKEVACSVIEIGGSFIE 562
Query: 642 FTSGGHHHPHKEAIQSRLEILIGQLKE 668
F +G + H H E IQ + +GQL +
Sbjct: 563 FAAGDYLHSHLEVIQ----LTLGQLSK 631
Score = 60.1 bits (144), Expect(2) = 2e-30
Identities = 27/63 (42%), Positives = 43/63 (67%), Gaps = 3/63 (4%)
Frame = +3
Query: 458 DLVSWNSMLKSYALHGKAKDALELFKKL---DVHPDSTTFVALLSACSHAGLVEEGVEIF 514
+ SWN+++ +A++G AK+ALE+F ++ P+ T + +LSAC+H GLVEEG F
Sbjct: 3 ETASWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSACNHCGLVEEGRRWF 182
Query: 515 NSM 517
N+M
Sbjct: 183 NAM 191
Score = 36.6 bits (83), Expect = 0.043
Identities = 19/53 (35%), Positives = 29/53 (53%), Gaps = 1/53 (1%)
Frame = +3
Query: 143 SWTALISGYAQCGLIRECFYLFSGLLAH-YRPNEFAFASLLSACEEHDIKCGM 194
SW ALI+G+A G +E +F+ ++ + PNE +LSAC CG+
Sbjct: 12 SWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSACNH----CGL 158
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.136 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,349,286
Number of Sequences: 63676
Number of extensions: 582830
Number of successful extensions: 3159
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 2886
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3055
length of query: 766
length of database: 12,639,632
effective HSP length: 105
effective length of query: 661
effective length of database: 5,953,652
effective search space: 3935363972
effective search space used: 3935363972
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0588c.4


