

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
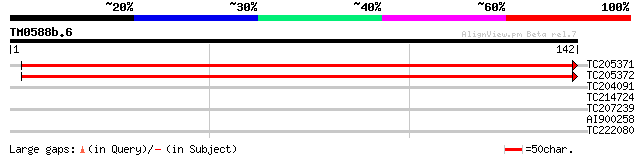
Query= TM0588b.6
(142 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205371 similar to UP|Q9FYG8 (Q9FYG8) F1N21.7, partial (43%) 222 4e-59
TC205372 similar to UP|Q9FYG8 (Q9FYG8) F1N21.7, partial (39%) 220 2e-58
TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1... 27 2.5
TC214724 homologue to UP|Q94FP3 (Q94FP3) Succinate dehydrogenase... 27 4.3
TC207239 similar to UP|Q9LTK0 (Q9LTK0) Arabidopsis thaliana geno... 25 9.5
AI900258 similar to SP|Q9SL48|SLY1 SEC1-family transport protein... 25 9.5
TC222080 weakly similar to GB|AAP37844.1|30725644|BT008485 At3g1... 25 9.5
>TC205371 similar to UP|Q9FYG8 (Q9FYG8) F1N21.7, partial (43%)
Length = 716
Score = 222 bits (566), Expect = 4e-59
Identities = 108/139 (77%), Positives = 120/139 (85%)
Frame = +2
Query: 4 AAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAA 63
A+K I HQIGG+QNDALRFGL GVKSDIVGSHPLQS+ +S +++E MKRQC VNLYG A
Sbjct: 86 ASKFIPHQIGGVQNDALRFGLHGVKSDIVGSHPLQSSLQSTRRMDEAMKRQCKVNLYGTA 265
Query: 64 FPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMH 123
FP+K +LDRQILSRFQRP G IPSSMLGLE TGS+D FGFED L+DPRESET RPLDMH
Sbjct: 266 FPMKEELDRQILSRFQRPPGPIPSSMLGLETVTGSVDHFGFEDCLNDPRESETFRPLDMH 445
Query: 124 HGMEVRLGLSKGPVCPSFM 142
HGMEVRLGLSKGPV PS +
Sbjct: 446 HGMEVRLGLSKGPVYPSII 502
>TC205372 similar to UP|Q9FYG8 (Q9FYG8) F1N21.7, partial (39%)
Length = 721
Score = 220 bits (561), Expect = 2e-58
Identities = 107/139 (76%), Positives = 119/139 (84%)
Frame = +3
Query: 4 AAKSIAHQIGGIQNDALRFGLQGVKSDIVGSHPLQSAQESASKINEVMKRQCMVNLYGAA 63
A+K I HQIGG+QND LRFGL GVKSDIVGSHPLQS+ +S +++E MK+QC VNLYG A
Sbjct: 78 ASKFIPHQIGGVQNDVLRFGLHGVKSDIVGSHPLQSSLQSTRRMDEAMKKQCKVNLYGTA 257
Query: 64 FPLKMDLDRQILSRFQRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMH 123
FPLK +LDRQILSRFQRP G IPSSMLGLE TGS+D FGFED L+DPRESET RPLDMH
Sbjct: 258 FPLKEELDRQILSRFQRPPGPIPSSMLGLETVTGSVDHFGFEDCLNDPRESETFRPLDMH 437
Query: 124 HGMEVRLGLSKGPVCPSFM 142
HGMEVRLGLSKGPV PS +
Sbjct: 438 HGMEVRLGLSKGPVYPSII 494
>TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(70%)
Length = 2075
Score = 27.3 bits (59), Expect = 2.5
Identities = 20/67 (29%), Positives = 30/67 (43%)
Frame = +2
Query: 36 PLQSAQESASKINEVMKRQCMVNLYGAAFPLKMDLDRQILSRFQRPSGAIPSSMLGLEAF 95
P+ + +S ++E +++ V+ GA DLD Q L F G I LGL+
Sbjct: 683 PMAPSAAPSSSVSEYTQKKIYVSNVGA------DLDPQKLLAFFSRFGEIEEGPLGLDKA 844
Query: 96 TGSLDDF 102
TG F
Sbjct: 845 TGKPKGF 865
>TC214724 homologue to UP|Q94FP3 (Q94FP3) Succinate dehydrogenase subunit 3
(Fragment), complete
Length = 1219
Score = 26.6 bits (57), Expect = 4.3
Identities = 25/105 (23%), Positives = 47/105 (43%), Gaps = 4/105 (3%)
Frame = +2
Query: 23 GLQGVKSDIVGS--HPLQSAQESASKI--NEVMKRQCMVNLYGAAFPLKMDLDRQILSRF 78
GL V ++ +GS + + S + AS + N + +C+ ++ A P+++ ++ + F
Sbjct: 302 GLPKVPANTLGSDSNAINSPRYKASGLDTNVLAGARCVSGVWAAQGPMRLVVNTVMARSF 481
Query: 79 QRPSGAIPSSMLGLEAFTGSLDDFGFEDYLSDPRESETIRPLDMH 123
+R A M+G + F + E LS RPL H
Sbjct: 482 ERSMEAGTLGMIGHKRFMSDIPSKTSETNLSG------FRPLSPH 598
>TC207239 similar to UP|Q9LTK0 (Q9LTK0) Arabidopsis thaliana genomic DNA,
chromosome 5, BAC clone:F17P19 (AT5g52200/F17P19_10),
partial (10%)
Length = 561
Score = 25.4 bits (54), Expect = 9.5
Identities = 11/32 (34%), Positives = 18/32 (55%)
Frame = +1
Query: 96 TGSLDDFGFEDYLSDPRESETIRPLDMHHGME 127
TGSL+D ED+ S P ++E + H ++
Sbjct: 151 TGSLEDESDEDHNSQPSKAEKCESSSLSHSVK 246
>AI900258 similar to SP|Q9SL48|SLY1 SEC1-family transport protein SLY1
(AtSLY1). [Mouse-ear cress] {Arabidopsis thaliana},
partial (20%)
Length = 426
Score = 25.4 bits (54), Expect = 9.5
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = -1
Query: 2 EEAAKSIAHQIGGIQNDALRFGLQGV 27
E +++ + GG+ NDAL GL G+
Sbjct: 375 EVEVEALVERFGGVGNDALHVGLVGL 298
>TC222080 weakly similar to GB|AAP37844.1|30725644|BT008485 At3g18210
{Arabidopsis thaliana;} , partial (25%)
Length = 688
Score = 25.4 bits (54), Expect = 9.5
Identities = 18/49 (36%), Positives = 22/49 (44%), Gaps = 13/49 (26%)
Frame = +3
Query: 99 LDDFGFEDYLSDPRESETIRP-------------LDMHHGMEVRLGLSK 134
LDDF FE L D + I+P LD HHG V G++K
Sbjct: 150 LDDFAFEAML-DRFMCDFIQPISRVFYPELGGSSLDSHHGFVVEYGINK 293
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,826,481
Number of Sequences: 63676
Number of extensions: 45992
Number of successful extensions: 193
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 193
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 193
length of query: 142
length of database: 12,639,632
effective HSP length: 88
effective length of query: 54
effective length of database: 7,036,144
effective search space: 379951776
effective search space used: 379951776
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0588b.6


