

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
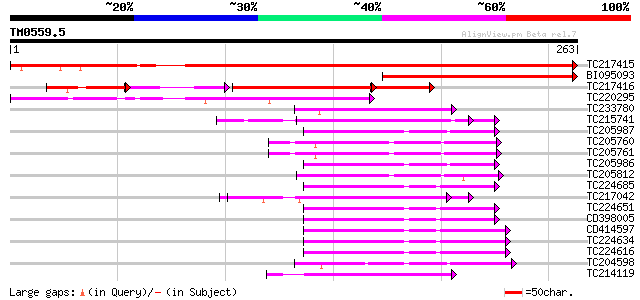
Query= TM0559.5
(263 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217415 similar to UP|O23166 (O23166) Thiol-disulfide interchan... 401 e-112
BI095093 similar to PIR|E85439|E854 thiol-disulfide interchange ... 169 1e-42
TC217416 similar to UP|O23166 (O23166) Thiol-disulfide interchan... 127 9e-39
TC220295 weakly similar to UP|O23166 (O23166) Thiol-disulfide in... 132 2e-31
TC233780 weakly similar to UP|Q9YKL1 (Q9YKL1) ORF1 protein, part... 56 2e-08
TC215741 UP|Q6I686 (Q6I686) Protein disufide isomerase-like prot... 55 3e-08
TC205987 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m... 55 4e-08
TC205760 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%) 54 6e-08
TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%) 54 7e-08
TC205986 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m... 54 9e-08
TC205812 similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, partial (64%) 52 2e-07
TC224685 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,... 52 3e-07
TC217042 UP|Q6I684 (Q6I684) Protein disulfide isomerase-like pro... 52 4e-07
TC224651 weakly similar to UP|THIM_BRANA (Q9XGS0) Thioredoxin M-... 51 6e-07
CD398005 51 6e-07
CD414597 50 1e-06
TC224634 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,... 50 1e-06
TC224616 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4,... 50 1e-06
TC204598 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%) 49 2e-06
TC214119 homologue to UP|Q6I687 (Q6I687) Protein disulfide isome... 49 2e-06
>TC217415 similar to UP|O23166 (O23166) Thiol-disulfide interchange like
protein (AT4g37200/C7A10_160) (Thioredoxin-like
protein), partial (66%)
Length = 1100
Score = 401 bits (1030), Expect = e-112
Identities = 206/266 (77%), Positives = 224/266 (83%), Gaps = 3/266 (1%)
Frame = +1
Query: 1 MSRL-SSNPIGLHRFRPCLRTPQ-FTVNPRHLH-FNTRKFHTLACQTNPNLDEKDASATS 57
M+RL SSNPI LHRF+PCL + F+ NPR NTRK TLACQTNPNLD+KD T
Sbjct: 67 MARLISSNPIALHRFQPCLPISRHFSANPRRRRPTNTRKLQTLACQTNPNLDQKDTPTTD 246
Query: 58 QEIKIVVEPSTVNGESETCKPTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRL 117
+E K+VVEPST + S LPQLPTK+IN K+A+ STLAALGLFL+ RL
Sbjct: 247 EE-KLVVEPST-------------STTTSSLPQLPTKNINNKIALVSTLAALGLFLSARL 384
Query: 118 DFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVM 177
DFGVSLKDL+ANA+PYE+ALSNGKPTVVEFYADWCEVCRELAPDVYK+EQQ+KDRVNFVM
Sbjct: 385 DFGVSLKDLSANAMPYEEALSNGKPTVVEFYADWCEVCRELAPDVYKVEQQFKDRVNFVM 564
Query: 178 LNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVP 237
LNVDNT WEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQ LLENVDALARGEASVP
Sbjct: 565 LNVDNTKWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQYLLENVDALARGEASVP 744
Query: 238 HARVVGKYSSAEARKVHQVVDPRSHG 263
HARVVG+YSSAEARKVHQV DPRSHG
Sbjct: 745 HARVVGQYSSAEARKVHQVADPRSHG 822
>BI095093 similar to PIR|E85439|E854 thiol-disulfide interchange like protein
[imported] - Arabidopsis thaliana, partial (34%)
Length = 457
Score = 169 bits (428), Expect = 1e-42
Identities = 81/90 (90%), Positives = 84/90 (93%)
Frame = -3
Query: 174 NFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGE 233
NFVMLNVDNT WEQELDEFG+EGIPHFAFLDK GNEEGNVVGRLPRQ LLENVDALARGE
Sbjct: 455 NFVMLNVDNTKWEQELDEFGMEGIPHFAFLDKIGNEEGNVVGRLPRQYLLENVDALARGE 276
Query: 234 ASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
SVPHARVVG+YS+AEARKVHQV DPR HG
Sbjct: 275 TSVPHARVVGQYSNAEARKVHQVADPRRHG 186
>TC217416 similar to UP|O23166 (O23166) Thiol-disulfide interchange like
protein (AT4g37200/C7A10_160) (Thioredoxin-like
protein), partial (35%)
Length = 827
Score = 127 bits (318), Expect(3) = 9e-39
Identities = 59/67 (88%), Positives = 65/67 (96%)
Frame = +3
Query: 104 STLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVY 163
STLAALGLFL+TRLDFGVSLKDL+A A+PYE+ALSNGKPTVVEFYADWCEVCRE APDVY
Sbjct: 363 STLAALGLFLSTRLDFGVSLKDLSAIAMPYEEALSNGKPTVVEFYADWCEVCREFAPDVY 542
Query: 164 KIEQQYK 170
K+EQQ+K
Sbjct: 543 KVEQQFK 563
Score = 55.8 bits (133), Expect = 2e-08
Identities = 24/30 (80%), Positives = 27/30 (90%)
Frame = +2
Query: 168 QYKDRVNFVMLNVDNTNWEQELDEFGVEGI 197
QY+D+VNFVMLNVDNT W ELDEFG+EGI
Sbjct: 725 QYRDQVNFVMLNVDNTKWGAELDEFGMEGI 814
Score = 39.7 bits (91), Expect(3) = 9e-39
Identities = 22/41 (53%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Frame = +2
Query: 18 LRTPQFTV--NPRHLHFNTRKFHTLACQTNPNLDEKDASAT 56
L+ P T+ N LH KF TLACQTNPNLD+KD T
Sbjct: 98 LQIPTITMPSNSSSLHC---KFQTLACQTNPNLDQKDTPTT 211
Score = 31.6 bits (70), Expect(3) = 9e-39
Identities = 18/49 (36%), Positives = 24/49 (48%)
Frame = +1
Query: 54 SATSQEIKIVVEPSTVNGESETCKPTSSTADASGLPQLPTKDINRKVAI 102
S+ E K+VVEPS + LPQLP KDIN K+++
Sbjct: 259 SSAQDEEKLVVEPSHTK---------------TSLPQLPAKDINNKISL 360
>TC220295 weakly similar to UP|O23166 (O23166) Thiol-disulfide interchange
like protein (AT4g37200/C7A10_160) (Thioredoxin-like
protein), partial (20%)
Length = 686
Score = 132 bits (332), Expect = 2e-31
Identities = 82/171 (47%), Positives = 103/171 (59%), Gaps = 2/171 (1%)
Frame = +2
Query: 1 MSRLSSNPIGLHRFRPCLRTPQFTVNPRHLHFNTRKFHTLACQTNPNLDEKDASATSQEI 60
M+ S N LH FRPC+ PQ NP L RK LACQ N NLD+ S+T++
Sbjct: 8 MACFSLNLSALHMFRPCVYIPQIVANP--LQHQNRKIKILACQANQNLDQ---SSTTE-- 166
Query: 61 KIVVEPSTVNGESETCKPTSSTADASGLP-QLPTKDINRKVAIASTLAALGLFLATRLDF 119
K V E G T+ +G+P + P+KD +K+AI S L ALGL L++RL F
Sbjct: 167 KSVAELGNSKG----------TSSPNGVPPKFPSKDFKKKIAIVSFLGALGLLLSSRLHF 316
Query: 120 -GVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQY 169
GV +KD A+ALP ++A NGK TVVEF ADWCEVC ELAPDV ++E QY
Sbjct: 317 FGVPMKDHCAHALPSKEARPNGKFTVVEFNADWCEVCGELAPDVSEVE*QY 469
>TC233780 weakly similar to UP|Q9YKL1 (Q9YKL1) ORF1 protein, partial (4%)
Length = 711
Score = 55.8 bits (133), Expect = 2e-08
Identities = 28/76 (36%), Positives = 43/76 (55%), Gaps = 1/76 (1%)
Frame = +2
Query: 133 YEQALSNGKP-TVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDE 191
+ AL++GK T+VEFY+ C +C L V ++E + + +N VM + +N NW EL
Sbjct: 227 FASALASGKEATLVEFYSPKCRLCNSLLKFVSEVETRNSNWLNIVMADAENPNWLPELLN 406
Query: 192 FGVEGIPHFAFLDKEG 207
+ V +P F LD G
Sbjct: 407 YDVSYVPCFVLLDNNG 454
>TC215741 UP|Q6I686 (Q6I686) Protein disufide isomerase-like protein,
complete
Length = 1703
Score = 55.1 bits (131), Expect = 3e-08
Identities = 37/131 (28%), Positives = 64/131 (48%)
Frame = +3
Query: 97 NRKVAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCR 156
+R++A+A+ AL LFL+ D V L + +E+ + + +VEFYA WC C+
Sbjct: 135 SRRIALAAFAFAL-LFLSASADDVVVLSEDN-----FEKEVGQDRGALVEFYAPWCGHCK 296
Query: 157 ELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGR 216
+LAP+ K+ +K + ++ VD + ++GV G P + K E G
Sbjct: 297 KLAPEYEKLGSSFKKAKSVLIGKVDCDEHKSLCSKYGVSGYPTIQWFPKGSLEPKKYEGP 476
Query: 217 LPRQLLLENVD 227
+ L+E V+
Sbjct: 477 RTAESLVEFVN 509
Score = 53.9 bits (128), Expect = 7e-08
Identities = 28/82 (34%), Positives = 40/82 (48%)
Frame = +3
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L K +VEFYA WC C+ LAP K+ +K + V+ N+D + +++
Sbjct: 585 EVVLDETKDVLVEFYAPWCGHCKSLAPTYEKVATAFKLEEDVVIANLDADKYRDLAEKYD 764
Query: 194 VEGIPHFAFLDKEGNEEGNVVG 215
V G P F K GN+ G G
Sbjct: 765 VSGFPTLKFFPK-GNKAGEDYG 827
>TC205987 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m {Pisum
sativum;} , partial (64%)
Length = 891
Score = 54.7 bits (130), Expect = 4e-08
Identities = 25/91 (27%), Positives = 51/91 (55%)
Frame = +1
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
+++ P +VEF+A WC CR +AP + ++ ++Y ++ LN D++ ++G+
Sbjct: 337 IASETPVLVEFWAPWCGPCRMIAPAIDELAKEYAGKIACFKLNTDDS--PNIATQYGIRS 510
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP F K G ++ +++G +P+ L V+
Sbjct: 511 IPTVLFF-KNGEKKESIIGAVPKSTLSATVE 600
>TC205760 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%)
Length = 666
Score = 54.3 bits (129), Expect = 6e-08
Identities = 31/109 (28%), Positives = 56/109 (50%), Gaps = 1/109 (0%)
Frame = +2
Query: 121 VSLKDLTANALPYEQALSNG-KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLN 179
V K T N+ ++ L+N KP +V+FYA WC C+ + P + ++ + KD++ V
Sbjct: 65 VQAKKQTYNS--FDDLLANSEKPVLVDFYATWCGPCQFMVPILNEVSTRLKDKIQVV--K 232
Query: 180 VDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDA 228
+D + D++ +E +P F + K+G G L L+E ++A
Sbjct: 233 IDTEKYPSIADKYRIEALPTF-IMFKDGEPYDRFEGALTADQLIERIEA 376
>TC205761 similar to UP|Q6NPF9 (Q6NPF9) At1g76760, partial (62%)
Length = 1023
Score = 53.9 bits (128), Expect = 7e-08
Identities = 31/109 (28%), Positives = 57/109 (51%), Gaps = 1/109 (0%)
Frame = +1
Query: 121 VSLKDLTANALPYEQALSNG-KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLN 179
V K T N+ +E L+N KP +V+FYA WC C+ + P + ++ + +D++ V
Sbjct: 379 VQAKKQTYNS--FEDLLANSEKPVLVDFYATWCGPCQFMVPILNEVSTRLQDKIQVV--K 546
Query: 180 VDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDA 228
+D + D++ +E +P F + K+G+ G L L+E ++A
Sbjct: 547 IDTEKYPTIADKYRIEALPTF-IMFKDGDPYDRFEGALTADQLIERIEA 690
>TC205986 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m {Pisum
sativum;} , partial (64%)
Length = 912
Score = 53.5 bits (127), Expect = 9e-08
Identities = 25/91 (27%), Positives = 50/91 (54%)
Frame = +1
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
+++ P +VEF+A WC CR +AP + ++ + Y ++ LN D++ ++G+
Sbjct: 349 IASETPVLVEFWAPWCGPCRMIAPVIDELAKDYAGKIACYKLNTDDS--PNIATQYGIRS 522
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP F K G ++ +++G +P+ L V+
Sbjct: 523 IPTVLFF-KNGEKKESIIGAVPKSTLSATVE 612
>TC205812 similar to UP|Q9SEU5 (Q9SEU5) Thioredoxin x, partial (64%)
Length = 904
Score = 52.4 bits (124), Expect = 2e-07
Identities = 32/98 (32%), Positives = 52/98 (52%), Gaps = 2/98 (2%)
Frame = +1
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
+ L +P +VEF A WC CR ++P + + ++Y+DR+ V +D+ + ++E+
Sbjct: 406 DTVLKANRPVLVEFVATWCGPCRLISPSMESLAKEYEDRLTVV--KIDHDANPRLIEEYK 579
Query: 194 VEGIPHFAFLDKEGNE--EGNVVGRLPRQLLLENVDAL 229
V G+P L K G E E G + + L E VDAL
Sbjct: 580 VYGLPTL-ILFKNGQEVPESRREGAITKVKLKEYVDAL 690
>TC224685 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (55%)
Length = 1169
Score = 52.0 bits (123), Expect = 3e-07
Identities = 25/91 (27%), Positives = 47/91 (51%)
Frame = +3
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L + P +VEF+A WC CR + P + ++ ++Y R+ LN D + +G+
Sbjct: 522 LESESPVLVEFWAPWCGPCRMIHPIIDELAKEYVGRLKCYKLNTDES--PSTATRYGIRS 695
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP + K G ++ V+G +P+ L +++
Sbjct: 696 IP-TVIIFKNGEKKDTVIGAVPKTTLTSSIE 785
>TC217042 UP|Q6I684 (Q6I684) Protein disulfide isomerase-like protein,
complete
Length = 1422
Score = 51.6 bits (122), Expect = 4e-07
Identities = 33/124 (26%), Positives = 51/124 (40%), Gaps = 6/124 (4%)
Frame = +1
Query: 98 RKVAIASTLAALGLFLATRLDFGVSLKDLTANALPY------EQALSNGKPTVVEFYADW 151
+K A T AL F+ V + + ++ + E K +VEFYA W
Sbjct: 457 KKYEGARTAEALAAFVNIEAGTNVKIASVASSVVVLSPNNFDEVVFDETKDVLVEFYAPW 636
Query: 152 CEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEG 211
C C+ LAP K+ + + V+ NVD ++ +++GV G P F K
Sbjct: 637 CGHCKALAPIYEKVAAAFNLDKDVVIANVDADKYKDLAEKYGVSGYPTLKFFPKSNKAGE 816
Query: 212 NVVG 215
N G
Sbjct: 817 NYDG 828
Score = 47.4 bits (111), Expect = 7e-06
Identities = 29/105 (27%), Positives = 51/105 (47%), Gaps = 1/105 (0%)
Frame = +1
Query: 102 IASTLAALGLFLATR-LDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAP 160
+A AL +FL++ D V+L + T +E + + +VEFYA WC C+ LAP
Sbjct: 142 LAIAAIALMMFLSSASADDVVALTEET-----FENEVGKDRAALVEFYAPWCGHCKRLAP 306
Query: 161 DVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDK 205
+ ++ +K + ++ VD + ++GV G P + K
Sbjct: 307 EYEQLGASFKKTKSVLIAKVDCDEHKSVCGKYGVSGYPTIQWFPK 441
>TC224651 weakly similar to UP|THIM_BRANA (Q9XGS0) Thioredoxin M-type,
chloroplast precursor (TRX-M), partial (67%)
Length = 848
Score = 50.8 bits (120), Expect = 6e-07
Identities = 23/91 (25%), Positives = 48/91 (52%)
Frame = +2
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
+ + P +VEF+A WC CR + P + ++ ++Y ++ LN D + ++G+
Sbjct: 338 IESESPVLVEFWAPWCGPCRMIHPIIDELAKEYTGKLKCYKLNTDES--PSTATKYGIRS 511
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP + K G ++ V+G +P+ L +++
Sbjct: 512 IP-TVIIFKNGEKKDTVIGAVPKTTLTSSIE 601
>CD398005
Length = 693
Score = 50.8 bits (120), Expect = 6e-07
Identities = 23/91 (25%), Positives = 48/91 (52%)
Frame = -3
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
+ + P +VEF+A WC CR + P + ++ ++Y ++ LN D + ++G+
Sbjct: 496 IESESPVLVEFWAPWCGPCRMIHPIIDELAKEYTGKLKCYKLNTDES--PSTATKYGIRS 323
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP + K G ++ V+G +P+ L +++
Sbjct: 322 IP-TVIIFKNGEKKDTVIGAVPKTTLTSSIE 233
>CD414597
Length = 663
Score = 49.7 bits (117), Expect = 1e-06
Identities = 25/96 (26%), Positives = 47/96 (48%)
Frame = -2
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L + +VEF+A WC CR + P + ++ +QY ++ LN D + +G+
Sbjct: 512 LESESAVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDES--PSTATRYGIRS 339
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARG 232
IP + K G ++ V+G +P+ L +++ G
Sbjct: 338 IP-TVMIFKNGEKKDTVIGAVPKSTLTASIEKFV*G 234
>TC224634 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (60%)
Length = 743
Score = 49.7 bits (117), Expect = 1e-06
Identities = 25/96 (26%), Positives = 47/96 (48%)
Frame = +2
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L + +VEF+A WC CR + P + ++ +QY ++ LN D + +G+
Sbjct: 296 LESESAVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDES--PSTATRYGIRS 469
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARG 232
IP + K G ++ V+G +P+ L +++ G
Sbjct: 470 IP-TVMIFKSGEKKDTVIGAVPKSTLTTSIEKFV*G 574
>TC224616 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (58%)
Length = 869
Score = 49.7 bits (117), Expect = 1e-06
Identities = 25/96 (26%), Positives = 47/96 (48%)
Frame = +3
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L + +VEF+A WC CR + P + ++ +QY ++ LN D + +G+
Sbjct: 366 LESESAVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDES--PSTATRYGIRS 539
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARG 232
IP + K G ++ V+G +P+ L +++ G
Sbjct: 540 IP-TVMIFKNGEKKDTVIGAVPKSTLTASIEKFV*G 644
>TC204598 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%)
Length = 610
Score = 48.9 bits (115), Expect = 2e-06
Identities = 35/107 (32%), Positives = 55/107 (50%), Gaps = 4/107 (3%)
Frame = +2
Query: 133 YEQALSNGKPT----VVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQE 188
+ Q L NGK + VV+F A WC CR +AP + +I ++ +V F+ ++VD
Sbjct: 101 WNQQLQNGKDSQKLIVVDFTASWCGPCRFIAPVLAEI-ARHTPQVIFLKVDVDEV--RPV 271
Query: 189 LDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEAS 235
+E+ +E +P F FL K+G VVG +L L ++ AS
Sbjct: 272 AEEYSIEAMPTFLFL-KDGKIVDKVVGAKKEELQLTIAKHVSAAAAS 409
>TC214119 homologue to UP|Q6I687 (Q6I687) Protein disulfide isomerase-like
protein, partial (30%)
Length = 770
Score = 48.9 bits (115), Expect = 2e-06
Identities = 26/88 (29%), Positives = 44/88 (49%)
Frame = +1
Query: 120 GVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLN 179
G SL+D+ +GK ++EFYA WC C++LAP + ++ Y++ + V+
Sbjct: 154 GASLEDIV---------FKSGKNVLLEFYAPWCGHCKQLAPILDEVAISYQNEADVVIAK 306
Query: 180 VDNTNWEQELDEFGVEGIPHFAFLDKEG 207
+D T + + F V+G P F G
Sbjct: 307 LDATANDIPSETFDVQGYPTVYFRSASG 390
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,664,762
Number of Sequences: 63676
Number of extensions: 134985
Number of successful extensions: 784
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 771
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 778
length of query: 263
length of database: 12,639,632
effective HSP length: 95
effective length of query: 168
effective length of database: 6,590,412
effective search space: 1107189216
effective search space used: 1107189216
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0559.5


